- Research article
- Open Access
- Published:
ANGUSTIFOLIAis a central component of tissue morphogenesis mediated by the atypical receptor-like kinase STRUBBELIG
BMC Plant Biologyvolume13, Article number:16(2013)
Abstract
Background
During plant tissue morphogenesis cells have to coordinate their behavior to allow the generation of the size, shape and cellular patterns that distinguish an organ. Despite impressive progress the underlying signaling pathways remain largely unexplored. InArabidopsis thaliana,atypical leucine-rich repeat receptor-like kinase STRUBBELIG (SUB) is involved in signal transduction in several developmental processes including the formation of carpels, petals, ovules and root hair patterning. The threeSTRUBBELIG-LIKE MUTANT(SLM) genesDETORQUEO(DOQ),QUIRKY(QKY) andZERZAUST(ZET) are considered central elements ofSUB-mediated signal transduction pathways as corresponding mutants share most phenotypic aspects withsub突变体s.
Results
Here we show thatDOQcorresponds to the previously identifiedANGUSTIFOLIAgene. The genetic analysis revealed that thedoq-1突变体exhibits all additional mutant phenotypes and conversely that otheranalleles show theslmphenotypes. We further provide evidence that SUB and AN physically interact and that AN is not required for subcellular localization of SUB.
Conclusions
Our data suggest thatANis involved inSUBsignal transduction pathways. In addition, they reveal previously unreported functions ofANin several biological processes, such as ovule development, cell morphogenesis in floral meristems, and root hair patterning. Finally, SUB and AN may directly interact at the plasma membrane to mediate SUB-dependent signaling.
Background
Tissue morphogenesis and cellular patterning require extensive cellular communication. In plants the coordination of cellular behavior within a tissue is intrinsically linked to cell wall biogenesis and dynamics, as plant cells are connected through semi-rigid cell walls that drastically limit their relative movement. It is a major current challenge in plant biology to understand the mechanistic basis of intercellular communication and its connection to the cell wall during tissue morphogenesis.
In Arabidopsis, intercellular signaling mediated by the atypical leucine-rich repeat transmembrane receptor-like (LRR-RLK) STRUBBELIG (SUB) is essential for a number of developmental processes [1–6]. Ovules ofsub突变体s show frequent defects in the initiation and outgrowth of the outer integument. In addition,sub突变体s exhibit twisted stems, petals and carpels/siliques. These phenotypes indicate a role forSUBin the control of integument initiation and outgrowth as well as stem and floral organ shape [1,2,6].SUBalso plays a role in internode length (and thus stem height), a trait that is potentially important for optimizing yield in crop plants.
At the cellular level, frequent misorientations of cell division planes were observed in e.g. L1 and L2 cells of young apical and floral meristems ofsub突变体s. Therefore, it was postulated thatSUBsignaling plays a role in orienting cell division planes in initiating integuments and floral meristems and thus influences the morphogenetic behavior of cells in a tissue context [1]. In addition,SUB, also known asSCRAMBLED(SCM), is involved in root hair patterning [3,4]. In this context,submutations lead to a randomization of root hair patterning such that root hairs develop ectopically or are not formed in the correct files.
In accordance with a perceived role ofSUBin coordinating cellular behavior in tissue morphogenesis and cell patterning,SUBacts in a non-cell-autonomous fashion and mediates inter-cell-layer signaling across histogenic cell layers in the ovule, the floral meristem [5] and the root [7].
SUB belongs to the LRRV/STRUBBELIG-RECEPTOR FAMILY (SRF) family [8,9] and has several protein domains including an extracellular domain with seven leucine-rich repeats, a transmembrane domain and a cytoplasmic putative kinase domain [1,3,6]. Interestingly, a set of biochemical and genetic data indicated that although the kinase domain is essential for SUB function, enzymatic phosphotransfer activity is not [1,6]. Thus, SUB is likely a so-called atypical or dead kinase.
Signaling by atypical kinases is poorly understood in plants [10,11]. In addition, a detailed structure-function analysis ofSUBsuggested that the organ or cell-specific aspects of SUB-mediated signaling are not integrated at the SUB receptor, but involve other components that act together with, or downstream of SUB [6]. In order to unravel the signal-transduction pathway ofSUBwe have previously identified three complementation groups sharing thesub-likemutant (slm) phenotypes [2]. In addition, it was found that there is significant overlap inSLM-sensitive gene expression. Taken together the results indicated thatSLMgenes contribute toSUBsignal transduction. The corresponding genes are calledQUIRKY(QKY),ZERZAUST(ZET), andDETORQUEO(DOQ) [2]. Initial molecular analysis suggested thatQKYencodes a putative membrane-localized protein with four C2 domains thus potentially connecting SUB to membrane-associated Ca2+- and phospholipid-dependent signaling [2].
In this work we focused on theDOQgene. We show thatdoq-1is a mutant allele of theANGUSTIFOLIA(AN) gene. Thedoq-1突变体carries a point mutation in theANgene and we further demonstrate thatdoq-1shares phenotypes with otheran等位基因和相反,其他analleles show allslmphenotypes tested. These results rule out the possibility thatdoq-1is an atypical allele. In addition, we provide evidence that SUB and AN can physically interact and thatANdoes not influence subcellular SUB distribution. Together our results reveal thatANis involved inSUB-dependent signaling events.
Results
doq-1突变体s exhibit an underbranched trichome and narrow leaf phenotype
Meiotic recombination mapping placedDOQin a 330.6 kb interval at the top of chromosome 1 (see Methods). This interval includedAN, a gene previously described to affect trichome branching, leaf morphology, and silique shape [12–15]. During the course of this analysis we noticed thatdoq-1trichomes are underbranched. Together with the previously described narrow leaf phenotype ofdoq-1[2], this suggested thatDOQandANfunctions are related. We therefore compared thedoq-1突变体leaf and trichome phenotype with three referenceanalleles,an-1,an-2,an-EM1和两个an-2 35S::YFP:ANrescued lines. Indoq-1突变体s, reductions in trichome branching approached levels seen in thean-1,an-2,an-EM1alleles (Figure1, Table1). Two-branched and three-branched trichomes are almost absent and a new class of unbranched trichomes was observed. The leaf shape ofdoq-1突变体s was also similar to that seen inan-1,an-2,an-EM1突变体s (Figure2). Correspondingly and in step with the referenceanalleles, the leaf length/width ratio indoq-1was significantly increased (Table2).
TheDOQgene corresponds to theANgene
We tested whether theDOQgene corresponds to theANgene. Genetic analysis revealed no complementation ofdoq-1withan-1indicating thatdoq-1is allelic toan. Furthermore, we sequenced theANgene in thedoq-1突变体and demonstrated a G to A transition at position 509 in the cDNA coding region. This mutation causes a glycine to aspartic acid substitution at position 170 that is located in the predicted NAD(P)-binding domain.
analleles showslmphenotypes
As mostslmphenotypes had not been reported foran突变体s, the question arose whetherdoq-1is an atypicalanallele showing new phenotypes or whether allanalleles share theslmphenotypes. We therefore comparedslmphenotypes betweendoq-1,an-1,an-2,an-EM1alleles andan-2 35S::YFP:ANrescued lines.
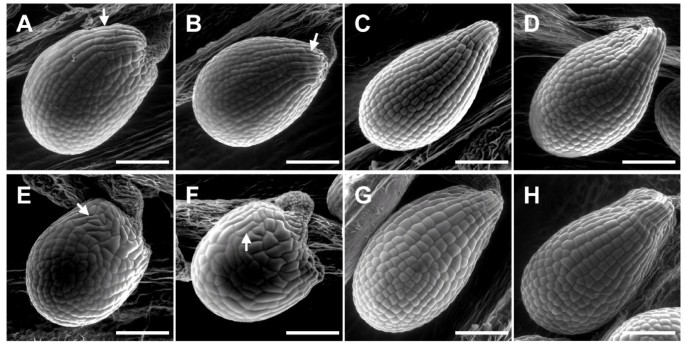
As described fordoq-1,allanalleles showed premature opening of flowers and twisted petals (Figure3). These phenotypes are rescued in thean-2 35S::YFP:ANlines. In addition,doq-1-like twisting of siliques was observed in allanalleles and could be rescued byANoverexpression (Figure4). Plants carrying thedoq-1mutation show a weak ovule phenotype as compared tosub突变体s [2]. To study the cellular patterns inanovules we used Scanning Electron Microscopy. We observed smaller cells with atypical division planes at the distal end of the outer integument in allanalleles tested and a rescue of this phenotype inan-2 35S::YFP:ANlines (Figure5). Finally, we analyzed the role ofANin root hair patterning. To this end we investigated the expression profiles of a GL2:GUS construct inan-1,an-2,an-EM1allelic backgrounds. GL2:GUS is expressed in atrichoblast cell files and serves as a convenient marker for root hair patterning [16]. Mutantan-1,an-2,an-EM1alleles displayed a severe distortion of the GL2:GUS pattern (Figure6). ThusANis a new component of the root hair patterning machinery.
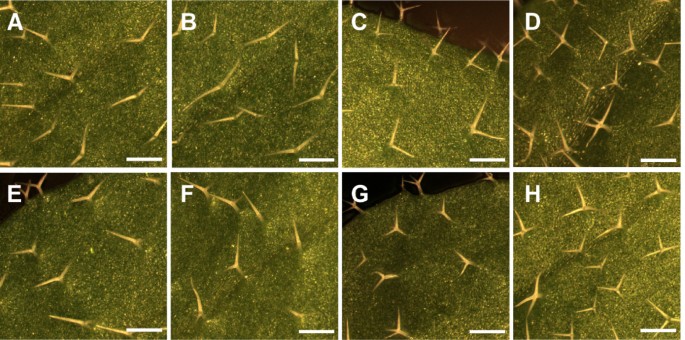
Inflorescence in WT,an突变体s and rescued lines.Prematurely opened flower buds in thean-1突变体(A), thean-2突变体(B), thedoq-1突变体(E) and thean-EM1突变体(F). Premature opening of flowers is rescued in thean-2 35S::YFP:AN#2 line (C) andan-2 35S::YFP:AN#4 line (D), as compared to WT Ler(G) and WT Col-0 (H). Scale bars: 3 mm.
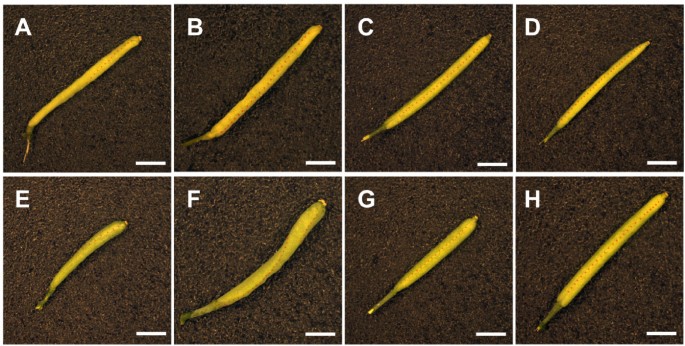
Silique morphology in WT,an突变体s and rescued lines.Twisted siliques are evident in thean-1突变体(A),an-2突变体(B),doq-1突变体(E) and thean-EM1突变体(F). Thean-2 35S::YFP:AN#2 line (C) andan-2 35S::YFP:AN#4 line (D) exhibit normal silique morphology, compared to WT Ler(G) and WT Col-0 (H). The replums are highlighted by dashed lines to indicate where twisting is present. Scale bars: 2 mm.
SUB and AN can interact directly
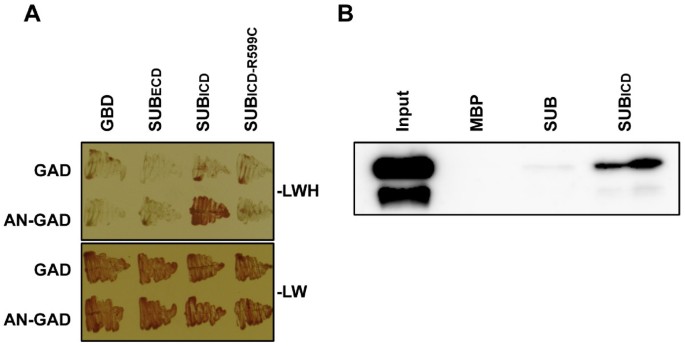
Next we addressed in more detail howSUBandDOQ/ANrelate to each other duringSUB-dependent signal transduction. Earlier results indicated that the two genes do not regulate each other at the transcriptional level [2]. We thus tested if DOQ/AN and SUB have the potential to interact directly at the protein level. Indeed, the intra-cellular domain of SUB (SUBICD, residues 371 to 768), but not the extra-cellular domain (SUBECD, residues 26 to 340), was able to interact with AN in a yeast two-hybrid assay (Figure7). In addition, fusions of maltose-binding protein (MBP) to SUBICD and the SUB full-length proteins were able to interact with a fusion of glutathione-S-transferase (GST) to AN in in vitro pull-down assays involving bacterially-expressed recombinant proteins (Figure7).
SUB can physically interact with AN.(A) Results of a yeast two-hybrid assay are depicted. The top panel shows that only cells carrying both plasmids encoding SUBICD and AN can grow on drop-out medium lacking leucine, tryptophan, and histidine (−LWH). Interaction depends on the presence of a functional SUBICD as indicated by the absence of signal in the variant SUBICD-R599C which mimics a strongsubmutation [6]. As a control all transformants can grow on medium lacking leucine and tryptophan (−LW) (bottom panel). Assays were done in the presence of 2.5 mM 3-amino-1,2,4-triazole (3-AT). GAD and GBD denote empty vector controls. (B) Western blot analysis of in vitro pull-down experiments to test MBP:SUB (full length) or MBP:SUBICD binding to GST:AN. AN was detected using a specific anti-AN antibody. The antibody detects two bands, both of which represent intact AN, as shown by MALDI-TOF analysis, suggesting two different conformations. GST:AN does not interact with MBP alone.
ANis not required for subcellular localization of SUB:EGFP in roots
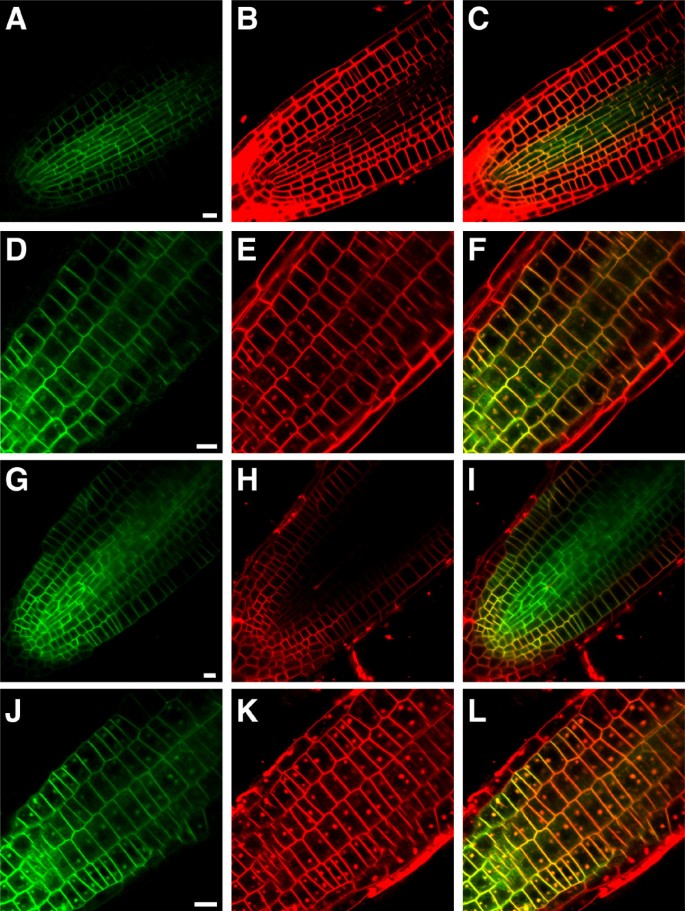
Previous studies involving a functional SUB:EGFP fusion protein indicated that SUB is localized to the plasma membrane and undergoes brefeldin A (BFA)-sensitive recycling [5–7]. As the AN homolog in animals was identified as a Brefeldin A ribosylated substrate [17] we tested whether the subcellular localization of SUB is dependent onDOQ/ANfunction. Towards this end we analyzed expression of a functionalSUB::SUB:EGFPreporter [6] in roots ofdoq-1突变体s in the absence or presence of BFA (Figure8). Expression of the reporter appeared normal under both experimental conditions indicating thatANis not required for correct subcellular localization and recycling of SUB.
SUB::SUB:EGFPexpression in five-days-old root tips of WT anddoq-1突变体s in the absence and presence of BFA.Confocal micrographs are shown. Two independent lines transgenic for a functionalSUB:SUB:EGFPreporter [6] exhibiting a GFP-based signal were analyzed for each genotype. GFP-based signal is shown in (A,D,G,J), FM4-64-based counterstain in (B,E,H,K) and the overlay in (C,F,I,L). Mid-optical section through a wild-type root tip highlights the plasma membrane localization of SUB:EGFP (A-C). Tangential optical section through the epidermis of a wild-type root tip treated with 50 μm BFA for 30 minutes reveals the typical dotted structures that indicate the presence of BFA compartments (D-F). A similar set is shown for untreateddoq-1roots (G-I) and BFA-treateddoq-1roots (J-L). Scale bars: 10 μm.
ERECTAinfluences theanphenotype
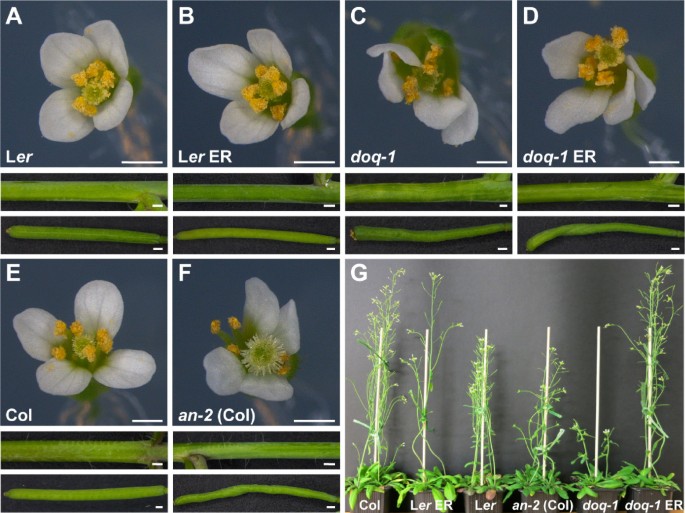
Thesubphenotype is sensitive to the genetic background [1] and it was shown that mutations inERECTA(ER), encoding a LRR-RLK [18], andSUBinteract synergistically assub erdouble mutants exhibit strongly reduced plant height [6]. To investigate this issue we transformed pKUT196, a plasmid containing 9.3 kb of Col-0 genomic DNA spanning the entireERlocus [18,19], intodoq-1(Ler) plants and asked whether a wild-type copy ofERwould affect thedoq-1phenotype. Interestingly, the transgene had a similar effect on plant height but not on other aspects of thedoq-1andsub-1phenotypes (Figure9) [6]. Similar tosub-1 ERplants, plant height was rescued indoq-1 ERplants indicating that height reduction indoq-1is caused by a synergistic interaction between thedoq-1andermutations. It should be noted, however, that plant height inan-2(Col) was slightly reduced as well, despite the presence of wild-typeER, though not to the extent as indoq-1(Ler). This suggests thatANaffects plant height in part independently ofER. Full rescue of plant height indoq-1 ERplants indicates the presence of additional modifiers that influence this trait in Ler.
Thedoq-1above-ground phenotype in the presence of functionalERECTA.Comparison of wild type,an-2(Col),doq-1(Ler) anddoq-1plants transgenic for ColER-containing plasmid pKUT196. (A-F) Morphology of flowers (upper panel), stems (central panel) and siliques (bottom panel). (A) Wild-type Ler. (B) Transgenic LerpKUT196. (C)doq-1突变体. Note the aberrant flower and silique morphology. Stem twisting is very mild if present at all. (D) Transgenicdoq-1pKUT196. Note the irregular flower and silique morphology. Stem morphology is essentially normal (compare with F). (E) Col wild type. (F) Colan-2突变体. Note the aberrant flower and silique morphology. Stem twisting is nearly normal. (G) Plant height comparisons of six-week-old pKUT196 transgenic plants in comparison to wild type and mutant reference lines. Note the height reduction inan-2(Col) plants. Scale bars: 0.5 mm.
Discussion
多个证据表明ANis involved in theSUB-dependent signaling mechanism. First, our genetic data show that all tested phenotypic aspects ofsubare shared byan突变体s. Second,DOQ/ANandSUBinfluence the expression of a common set of target genes. For example, 62 percent of genes misexpressed insubflowers are also misexpressed indoq-1/an[2]. Thirdly, AN and SUB are able to interact physically in two different assays. Finally,anandermutations synergistically affect internode length and plant height, as was observed forsubander[6]. These results unexpectedly bring together two well-established but previously unconnected research fields. What do we learn for the function ofANand what for the function ofSUB?
ANis involved in a broad variety of cellular processes
ANwas initially identified by its narrow leaf phenotype and trichome phenotype [14]. The closer analysis of the narrow leaf phenotypes suggested a role in cell polarity of leaf cells, such that the length of individual cells was increased and the width reduced [15,20]. As cell number is also changed an additional role in cell division control was postulated. The characterization of the trichome phenotype placedANin a regulatory network controlling branching initiation [12,13]. In this context, a conspicuous lack of microtubule accumulation in the branch initiation zone of the developing trichome cell suggested a role of AN in the control of microtubule organization [21]. The newslmphenotypes ofan突变体s reported in this study indicate thatANis involved in aSUBdependent signal transduction cascade. This function is separate from the first two functional aspects derived from leaf shape and trichome phenotypes, assub突变体s do not share these phenotypic aspects [2]. Such a broad spectrum of functions is compatible with the proposed biochemical functions of the AN protein. TheANgene encodes a protein with homology to CtBP/BARS [21,22]. CtBPs (C-terminal Binding Protein) were initially discovered as proteins binding to the C-terminal domain of adenovirus EA-1 [23]. CtBPs have been described as co-repressors for many transcriptional repressors carrying a PxDLS or RRT protein motif [24,25]. A possible function of AN as a co-repressor is the finding that micro-array experiments revealed many genes that are transcriptionally regulated by AN [2,22]. CtBP/BARS were independently identified as Brefeldin A ADP ribosylated substrates (BARS) [17]. A possible Golgi-related function is supported by the finding that CtBP can induce constriction in Golgi tubules [26] and membrane fission [27]. In support of such a function AN was reported to act outside the nucleus [28].
Role ofANinSUB-mediated signal transduction
How doesANfit into theSUB-mediated signaling pathway?ANis unlikely to be a direct or indirect transcriptional target ofSUBsignaling. For example,SUBexpression is only minimally altered indoq-1flowers at various stages [2]. In addition, a35S::SUBtransgene failed to rescue the phenotype ofdoq-1突变体s indicating thatSUBis not directly regulated at the transcriptional level byAN. At the same time,ANexpression was not found to differ between floral tissue of wild type,sub-1and otherslm突变体s [2] (data not shown). These observations render it unlikely thatANandSUBregulate each other’s activity at the transcriptional level.
Subcellular localization of a functional SUB:EGFP fusion protein was found to be restricted to the plasma membrane [6,7]. A functional AN:GFP fusion protein was recently reported to reside in the cytoplasm and in punctate compartments around the trans-Golgi network (TGN) [28]. The TGN localization of AN led the authors to suggest a Golgi-related role for this protein, possibly in membrane trafficking. These results are compatible with at least two possibilities of how AN may fit into the SUB signaling mechanism. In the first scenario AN could mediate membrane trafficking of SUB, a view that is also indirectly supported by the finding thatQKYencodes a putative membrane-localized protein thought to function in membrane-associated Ca2+- and phospholipid-dependent signaling [2]. However, this model does not fit the data presented in this study as signal distribution of a functionalSUB::SUB:EGFPreporter was found to be unaltered indoq-1roots. In an alternative scenario, the cytoplasmic distribution of AN would allow its direct interaction with the intracellular domain of SUB.
我们目前支持这个概念的结果yeast two-hybrid and in vitro pull-down assays suggest direct interaction between SUB and AN proteins at the plasma membrane. This interaction could then be necessary to control further downstream events of SUB signal transduction. AN is likely to mediate only some aspects of SUB signaling as there is a difference in the strength between for example the ovule phenotypes ofsubanddoq-1突变体s and stem twisting is nearly absent indoq-1oran-2(Figure9). Moreover, there is only partial overlap in misexpressed genes betweensubanddoq-1flowers [2]. With respect to plant height AN appears to be part of the SUB mechanism that interacts with the LRR-RLK ER. It was proposed thatERandSUBsignaling converge either at the level of the receptors or at some level more downstream in the mechanism [6]. Thus, it will be interesting to resolve exactly how SUB, AN and ER relate to each other in future studies on SUB signaling.
Conclusions
In this study we showed that phenotypes of theslm突变体doq-1and the trichome and leaf shape mutantanoverlap. In addition, we showed thatdoq-1is allelic toan. We further demonstrated thatdoq-1is not an atypicalanallele but that all testedan突变体s show previously undescribedslmaspects. The data reveal a broader range of biological functions forANthan previously appreciated. Finally, our data reveal the possibility that SUB and AN interact directly. Taken together, the presented evidence suggests a role for AN in tissue morphogenesis mediated by the atypical receptor-like kinase SUB.
Methods
Plant work and genetics
The followingArabidopsis thaliana(L.) Heynh. lines were used in this study: Columbia (Col-0) and Landsberg (erecta突变体) (Ler) wild-type strains,doq-1[2],an-1,an-2 [22],an-EM1[21],an-2 35S::YFP:AN.an-2 35S::YFP:ANplants were generated by cloning the CDS of the YFP:AN fusion into the pPAMPAT vector containing the 35S promoter (GenBank accession AY027531). Plasmid pKUT196 was described previously [19]. Plant transformation was performed according to the floral dip method [29].
植物生长在土壤在24°C的16个小时light per day. TheGL2:GUSline (Ler) [16] was introduced intoan突变体s by backcrosses. For GUS assays, plants were grown on MS plates for 4 days.
Using a Ler/Col mapping populationdoq-1was localized to the upper end of chromosome 1 between markers F10O3(481D) and NF21M12 [2]. Further mapping revealed an interval of 330.6 kb. The final Northern marker 96_(BccI) was located at chromosomal position 96771 (one recombinant left). 96_(BccI) is a CAPS marker which yields the following products after PCR with primers 96_(BccI)_F (GGGCTTTGATTTGATTGTGG) and 96_(BccI)_R (AAGAGAGGAGTGCAGCCAAA) and BccI digestion: Ler - 498 bp, Col - 254, 244 bp. The final Southern marker was NT7123 (chromosomal position: 427,343 bp, 3 recombinants left). NT7123 is a SSLP marker that, following PCR with primers NT7123_F (GTGTCCTTTTTTCTCAACGATG) and NT7123_R (CATGCACGTACGATTTGTTTAAC), yielded the following products on a 3.5% agarose gel: Ler , <199 bp; Col, 199 bp.
Yeast two-hybrid assay
The Matchmaker yeast two-hybrid system (Clontech) was employed and experimental procedures followed the manufacturer’s recommendations. The pACT-AN construct was described previously [21]. For the generation of pGBKT7-SUBICD, the intracellular part of SUB coding sequence [1] was amplified from cDNA using primers SUBintra_F (CATGCCATGGATAACCGATATTACAGTG) and SUBintra_R (ATCGGTCGACAATAAACTATTGCTTCTG). The PCR product was digested with NcoI/SalI and cloned into NcoI/SalI digested pGBKT7. The R599C mutation was introduced into pGBKT7-SUBICD by the QuikChange II XL site-directed mutagenesis kit according to the manufacturer’s recommendations (Agilent Technologies) by using primers 35SsubR599Cf (AAGAAGCTCACTTGGAATGTATGTATAAATATTGCATTAGGAGCTTC) and 35SsubR599Cr (GAAGCTCCTAATGCAATATTTATACATACATTCCAAGTGAGCTTCTT). For the cloning of pGBKT7-SUBECD, the extracellular part of SUB coding sequence was amplified from cDNA using primers SUB Extra/Nde1_F (GCTCATATGACTAATCTACGAGATGTTTCGGCGA and SUB Extra/Xma1_R (TACCCGGGGTTGAGTGGACCAGAATTTTCCTGATC). The PCR product was digested with NdeI/XmaI and cloned into NdeI/XmaI digested pGBKT7. All PCR-based constructs were sequenced.
To assay possible interactions in yeast pGBKT7 plasmids containing SUBECD, SUBICD and SUBICD-R599C were cotransformed with pACT or pACT-AN into yeast strain AH109. Transformants were selected after 3 days on SC medium lacking Leu and Trp (−LW) at 30°C. To examine yeast two-hybrid interactions, the transformants were grown on solid SC medium lacking Leu and Trp (SC-LW) or Leu, Trp, and His (SC-LWH) at 30°C.
Generation of constructs for recombinant protein production
ANandSUBcDNA were cloned into gateway entry vector pDONR 201. The following primers were used for gateway cloning: for amplifyingANcDNA: GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGAGCAAGATCCGTTCG and GGGGACCACTTTGTACAAGAAAGCTGGGTCTTAATCGATCCAACGTGTGATAC; for amplification of the full lengthSUBcDNA: GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGAGCT TTACAAGATGGGAAGTGT and GGGGACCACTTTGTACAAGAAAGCTGGGTCTTAGATCATATGTTGA AGATCTTGG; for the amplification of the ICD version of SUB: GGGGACAAGTTTGTACAAAAAAGCAGGCTTCatgTATAACCGATATTACAGTGGAGC and GGGGACCACTTTGTACAAGAAAGCTGGGTCTTAGATCAT ATGTTGAAGATCTTGG.ANcDNA was cloned into pGEX2TMGW (GE Healthcare) to generate an N-terminal fusion with Glutathion-S-Transferase (GST:AN) and the SUB and SUBICD cDNAs were cloned into pETG-40a (EMBL, Heidelberg) to generate an N-terminal fusion with maltose-binding protein (MBP:SUB, MBP:SUBICD).
Antibody generation
To produce anti-AN antibody, AN was expressed as a GST-fusion protein inE.coli. The protein was purified and used to generate antibodies in rabbit (Pineda Antikörper-Service; Berlin, Germany). The antibody serum was affinity purified and checked for its specificity by MALDI-TOF analysis.
In vitro pull-down assay
子之间的相互作用和使用进行了研究purified proteins that were expressed in bacteria. The bacterial cells BL21-CodonPlus (DE3)-RIL containing the IPTG inducible constructs (MBP-Full length SUB, MBP-SUBICD and GST-AN) were grown at 37°C and 220 rpm to an OD 600 of 0.8-0.9 and then the cultures were induced by adding IPTG to final concentration of 1 mM. The induced cells were then grown further for 5 hours (at 37°C for GST-tagged AN constructs and 20°C for MBP-tagged constructs) and cells were harvested by centrifugation. Cells were lysed in Tris-lysis buffer (Tris (pH 7.5) 50 mM, NaCl (100 mM), EDTA (1 mM), EGTA (1 mM), NP-40 (1%), Lysozyme (200 μg/ml), DTT (1 mM), Protease inhibitor cocktail (Sigma)) and sonicated three times for one minute each. The supernatant was collected by centrifugation at 4°C. MBP-tagged proteins were purified by incubation with amylose resin overnight at 4°C. After several washings, part of the resin was boiled with SDS-PAGE gel loading buffer to get purified protein for analysis. The remaining resin was used for incubation with equal amounts of GST:AN lysate for 4 hours at 4°C followed by several washings. Finally, the beads were boiled in SDS-PAGE gel loading buffer at 96°C for 10 min and equal amounts were loaded on a gel followed by western blotting. Detection was done using primary anti-AN antibody and secondary anti-rabbit antibody using SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific).
Histochemical analysis, microscopy and BFA treatments
Histochemical localization of ß-glucuronidase (GUS) activity in whole-mount tissues was performed as described previously [16]. Scanning Electron Microscopy was made using a Quanta 250 FEG (FEI) microscope under low vacuum conditions without any fixation steps. Confocal laser scanning microscopy and BFA treatments were performed as reported previously [6].
References
Chevalier D, Batoux M, Fulton L, Pfister K, Yadav RK, Schellenberg M, Schneitz K:STRUBBELIGdefines a receptor kinase-mediated signaling pathway regulating organ development inArabidopsis. Proc Natl Acad Sci USA. 2005, 102: 9074-9079. 10.1073/pnas.0503526102.
Fulton L, Batoux M, Vaddepalli P, Yadav RK, Busch W, Andersen SU, Jeong S, Lohmann JU, Schneitz K:DETORQUEO,QUIRKY, andZERZAUSTrepresent novel components involved in organ development mediated by the receptor-like kinase STRUBBELIG inArabidopsis thaliana. PLoS Genet. 2009, 5: e1000355-10.1371/journal.pgen.1000355.
Kwak SH, Shen R, Schiefelbein J: Positional signaling mediated by a receptor-like kinase in Arabidopsis. Science. 2005, 307: 1111-1113. 10.1126/science.1105373.
Kwak SH, Schiefelbein J: The role of the SCRAMBLED receptor-like kinase in patterning theArabidopsisroot epidermis. Dev Biol. 2007, 302: 118-131. 10.1016/j.ydbio.2006.09.009.
Yadav RK, Fulton L, Batoux M, Schneitz K: TheArabidopsisreceptor-like kinase STRUBBELIG mediates inter-cell-layer signaling during floral development. Dev Biol. 2008, 323: 261-270. 10.1016/j.ydbio.2008.08.010.
Vaddepalli P, Fulton L, Batoux M, Yadav RK, Schneitz K: Structure-function analysis of STRUBBELIG, an Arabidopsis atypical receptor-like kinase involved in tissue morphogenesis. PLoS One. 2011, 6: e19730-10.1371/journal.pone.0019730.
Kwak SH, Schiefelbein J: A feedback mechanism controlling SCRAMBLED receptor accumulation and cell-type pattern inArabidopsis. Curr Biol. 2008, 18: 1949-1954. 10.1016/j.cub.2008.10.064.
Shiu S-H, Bleecker AB: Receptor-like kinases fromArabidopsisform a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA. 2001, 98: 10763-10768. 10.1073/pnas.181141598.
Eyüboglu B, Pfister K, Haberer G, Chevalier D, Fuchs A, Mayer KF, Schneitz K: Molecular characterisation of theSTRUBBELIG-RECEPTOR FAMILYof genes encoding putative leucine-rich repeat receptor-like kinases inArabidopsis thaliana. BMC Plant Biol. 2007, 7: 16-10.1186/1471-2229-7-16.
Castells E, Casacuberta JM: Signalling through kinase-defective domains: the prevalence of atypical receptor-like kinases in plants. J Exp Bot. 2007, 58: 3503-3511. 10.1093/jxb/erm226.
吉斯拉,克拉克SE: RLK /皮尔激酶家族. Plant J. 2011, 66: 117-127. 10.1111/j.1365-313X.2011.04518.x.
Folkers U, Berger J, Hülskamp M: Cell morphogenesis of trichomes inArabidopsis: differential control of primary and secondary branching by branch initiation regulators and cell growth. Development. 1997, 124: 3779-3786.
Hülskamp M, Misera S, Jürgens G: Genetic dissection of trichome cell development in Arabidopsis. Cell. 1994, 76: 555-566. 10.1016/0092-8674(94)90118-X.
Redei GP: Single locus heterosis. Z Vererbungsl. 1962, 93: 164-170.
Tsuge T, Tsukaya H, Uchimiya H: Two independent and polarized processes of cell elongation regulate leaf blade expansion inArabidopsis thaliana(L.) Heynh. Development. 1996, 122: 1589-1600.
Masucci JD, Rerie WG, Foreman DR, Zhang M, Galway ME, Marks MD, Schiefelbein JW: The homeobox geneGLABRA2is required for position-dependent cell differentiation in the root epidermis ofArabidopsis thaliana. Development. 1996, 122: 1253-1260.
斯帕诺年代,Silletta毫克,Colanzi阿尔贝蒂年代,FiucciG, Valente C, Fusella A, Salmona M, Mironov A, Luini A, Corda D: Molecular cloning and functional characterization of brefeldin A-ADP-ribosylated substrate. A novel protein involved in the maintenance of the Golgi structure. J Biol Chem. 1999, 274: 17705-17710. 10.1074/jbc.274.25.17705.
Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y: The ArabidopsisERECTAgene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell. 1996, 8: 735-746.
Godiard L, Sauviac L, Torii KU, Grenon O, Mangin B, Grimsley NH, Marco Y: ERECTA, an LRR receptor-like kinase protein controlling development pleiotropically affects resistance to bacterial wilt. Plant J. 2003, 36: 353-365. 10.1046/j.1365-313X.2003.01877.x.
Tsukaya H, Tsuge T, Uchimiya H: The cotyledon: a superior system for studies of leaf development. Planta. 1994, 195: 309-312.
Folkers U, Kirik V, Schobinger U, Falk S, Krishnakumar S, Pollock MA, Oppenheimer DG, Day I, Reddy AS, Jürgens G, Hülskamp M: The cell morphogenesis geneANGUSTIFOLIAencodes a CtBP/BARS-like protein and is involved in the control of the microtubule cytoskeleton. EMBO J. 2002, 21: 1280-1288. 10.1093/emboj/21.6.1280.
Kim GT, Shoda K, Tsuge T, Cho KH, Uchimiya H, Yokoyama R, Nishitani K, Tsukaya H: TheANGUSTIFOLIAgene ofArabidopsis, a plantCtBPgene, regulates leaf-cell expansion, the arrangement of cortical microtubules in leaf cells and expression of a gene involved in cell-wall formation. EMBO J. 2002, 21: 1267-1279. 10.1093/emboj/21.6.1267.
Boyd JM, Subramanian T, Schaeper U, La Regina M, Bayley S, Chinnadurai G: A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. EMBO J. 1993, 12: 469-478.
Chinnadurai G: CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol Cell. 2002, 9: 213-224. 10.1016/S1097-2765(02)00443-4.
Quinlan KG, Nardini M, Verger A, Francescato P, Yaswen P, Corda D, Bolognesi M, Crossley M: Specific recognition of ZNF217 and other zinc finger proteins at a surface groove of C-terminal binding proteins. Mol Cell Biol. 2006, 26: 8159-8172. 10.1128/MCB.00680-06.
Weigert R, Silletta MG, Spano S, Turacchio G, Cericola C, Colanzi A, Senatore S, Mancini R, Polishchuk EV, Salmona M, Facchiano F, Burger KN, Mironov A, Luini A, Corda D: CtBP/BARS induces fission of Golgi membranes by acylating lysophosphatidic acid. Nature. 1999, 402: 429-433. 10.1038/46587.
Bonazzi M,斯帕诺年代,Turacchio G, Cericola C,瓦伦te C, Colanzi A, Kweon HS, Hsu VW, Polishchuck EV, Polishchuck RS, Sallese M, Pulvirenti T, Corda D, Luini A: CtBP3/BARS drives membrane fission in dynamin-independent transport pathways. Nat Cell Biol. 2005, 7: 570-580. 10.1038/ncb1260.
佐藤Minamisawa N, M,赵KH,上野H, Takechi K, Kajikawa M, Yamato KT, Ohyama K, Toyooka K, Kim GT, Horiguchi G, Takano H, Ueda T, Tsukaya H: ANGUSTIFOLIA, a plant homolog of CtBP/BARS, functions outside the nucleus. Plant J. 2011, 68: 788-799. 10.1111/j.1365-313X.2011.04731.x.
Clough SJ, Bent AF: Floral dip: a simplified method forAgrobacterium-mediated transformation ofArabidopsis thaliana. Plant J. 1998, 16: 735-743. 10.1046/j.1365-313x.1998.00343.x.
Acknowledgements
We thank B. Ülker (MPIZ, Cologne) for providing the pAMPAT-GW vector and K. Torii for the pKUT196 plasmid. We also thank members of the Hülskamp and Schneitz labs for stimulating discussions. This work was funded through grants SCHN 723/6-1 and SFB924 (TP A2) from the German Research Council (DFG) to KS. YB was funded by the International Max Planck Research School “Molecular Basis of Plant Development and Environmental Interaction”. HB was funded by an IGSDHD fellowship.
Author information
Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
LF, MH, KS designed the study. YB, LF, PV and HB performed experiments. YB, LF, PV, HB, MH and KS analyzed the data. MH and KS wrote the manuscript. All authors read and approved the final manuscript.
Yang Bai, Prasad Vaddepalli, Lynette Fulton, Hemal Bhasin contributed equally to this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open AccessThis article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Bai, Y., Vaddepalli, P., Fulton, L.et al.ANGUSTIFOLIAis a central component of tissue morphogenesis mediated by the atypical receptor-like kinase STRUBBELIG.BMC Plant Biol13,16 (2013). https://doi.org/10.1186/1471-2229-13-16
Received:
Accepted:
Published:
DOI:https://doi.org/10.1186/1471-2229-13-16
Keywords
- ANGUSTIFOLIA
- Flower
- Ovule
- Receptor-like kinase
- Root hair patterning
- Signal transduction
- Tissue morphogenesis
- STRUBBELIG