- Research article
- Open Access
- Published:
High-throughput genotyping of wheat-barley amphiploids utilising diversity array technology (DArT)
BMC Plant Biologyvolume13, Article number:87(2013)
Abstract
Background
Hordeum chilense, a native South American diploid wild barley, is one of the species of the genusHordeumwith a high potential for cereal breeding purposes, given its high crossability with other members of the Triticeae tribe. Hexaploid tritordeum (×TritordeumAscherson et Graebner, 2n=6×=42, AABBHchHch) is the fertile amphiploid obtained after chromosome doubling of hybrids betweenHordeum chilenseand durum wheat. Approaches used in the improvement of this crop have included crosses with hexaploid wheat to promote D/Hchchromosome substitutions. While this approach has been successful as was the case with triticale, it has also complicated the genetic composition of the breeding materials. Until now tritordeum lines were analyzed based on molecular cytogenetic techniques and screening with a small set of DNA markers. However, the recent development of DArT markers inH. chilenseoffers new possibilities to screen large number of accessions more efficiently.
Results
Here, we have applied DArT markers to genotype composition in forty-six accessions of hexaploid tritordeum originating from different stages of tritordeum breeding program and toH. chilense-wheat chromosome addition lines to allow their physical mapping. Diversity analyses were conducted including dendrogram construction, principal component analysis and structure inference. Euploid and substituted tritordeums were clearly discriminated independently of the method used. However, dendrogram and Structure analyses allowed the clearest discrimination among substituted tritordeums. The physically mapped markers allowed identifying these groups as substituted tritordeums carrying the following disomic substitutions (DS): DS1D (1Hch), DS2D (2Hch), DS5D (5Hch), DS6D (6Hch) and the double substitution DS2D (2Hch), DS5D (5Hch).These results were validated using chromosome specific EST and SSR markers and GISH analysis.
Conclusion
In conclusion, DArT markers have proved to be very useful to detect chromosome substitutions in the tritordeum breeding program and thus they are expected to be equally useful to detect translocations both in the tritordeum breeding program and in the transference ofH. chilensegenetic material in wheat breeding programs.
Background
Interspecific and intergeneric hybridization is a useful tool in the breeding of cultivated species of Triticeae tribe. This technique has been widely used to transfer desirable traits from wild to cultivated species [1–3] and to increase the genetic variation of the species by developing new synthetic hexaploid wheats [4–6] or usingTriticum urartu(donor of the A genome) for durum wheat breeding [7].Intergeneric hybrids betweenHordeumandTriticumgenus were attempted since the beginning of the 20thcentury. Since the first hybrids reported by [8] numerous hybrid combinations between both genera have been produced and reviewed by [9].However, only a few fertile hybrids have been obtained by chromosome doubling with colchicine. These were the hybrids betweenT. timopheevii×H. bogdanii[10],H. chilense×T. aestivum[11] andH. chilense×T. durum[12].AmongHordeumspecies,Hordeum chilenseRoem. et Schultz. is a native South American diploid perennial wild barley (2n = 2× = 14), included in the section Anisolepsis [13].It belongs to a heterogeneous group of South AmericanHordeumspecies and it is one of the species of the genusHordeumwith a high potential for cereal breeding purposes, given its high crossability with other members of the Triticeae tribe and other interesting characteristics [14].近年来,分子和细胞学techniques have been developed inH. chilensefor basic cytogenetic research, genetic diversity studies and monitoringH. chilensechromosomes in wheat genetic background. Several types of molecular markers, including Random Amplified Polymorphic DNA (RAPD) [15], Sequence Characterized Amplified Regions (SCARs) [16], Cleaved Amplified Polymorphisms (CAPs) [17] or DArTs [18] have been developedde novoforH. chilense. Similarly other markers have been transferred from wheat and barley species including Sequence Tagged Sites (STSs) [19], genomic or EST-derived simple sequence repeat (SSRs) [20,21], ESTs [22–24] or Conserved Orthologus Sequences (COS) [18].
In addition to marker-assisted selection, genomic in situ hybridization (GISH) and fluorescence in situ hybridization (FISH) are useful tools which have been used for identifyingH. chilense在小麦染色体或染色体易位background [25–27].
Tritordeums (×TritordeumAscherson et Graebner) are the fertile amphiploids obtained after chromosome doubling of hybrids between wheat (Triticumsp.) andH. chilense. They have been synthesised at different ploidy levels and genome constitutions, of which hexaploids tritordeums (2n = 6× = 42, AABBHchHch) have been subject of breeding program [28].The favourable agronomic traits shown by tritordeums, such as high biomass yield, number of spikelets/spike, seed size and high protein content, suggested its potential to become a new crop [29] which was confirmed nearly two decades later [30].The high seed carotenoid content [31,32] of this species constitutes also an interesting characteristic in the context of developing functional foods. Accordingly, tritordeum has received attention as a potential crop in the last years and a breeding program has been developed including the development of chromosome substitutions involving D and Hchgenomes. While the chromosome substitution program has obtained successful results for free threshing ability [33烘焙面包)或提高质量(34], it has also complicated the genetic composition of breeding materials since different chromosome substitutions may be present involving D and Hchgenomes. Until now tritordeum lines were inspected forH. chilense/wheat substitutions using physically mapped markers per chromosome [33] and using cytogenetics tools like GISH [22,24] and FISH [35].While cytogenetics is useful for a limited set of lines, it is impractical for large amounts of entries included in a breeding program. Furthermore, the use of a limited number of molecular markers is not adequate for managing a large collection of germplasm since the rate of error in classification may be too high compared to whole genome profiling.
Based on the above considerations we embarked on development of Diversity Arrays Technology (DArT) since this technology can overcome these constraints. In addition DArT technology offers low cost per data, high throughput and sequence-independent genotyping [36,37] and allows simultaneously determination of several hundred to several thousands of polymorphic loci spread over the genome [38].The high number of DArT markers generated in a single assay not only provides a precise estimate of genetic relationships among genotypes, but also their distribution over the genome offers real advantages for a range of molecular breeding and genomics applications.
DArT markers have been developed in more than 70 species (http://www.diversityarrays.com), including cereals such as cultivated barley (Hordeum vulgare), wheat (Triticum aestivum), durum wheat (Triticum durum), and recently wild barley (H. chilense).
DArT array have also been proved useful for polyploidy species with highly complex genomes [39]; to provide a fast and accurate means of determining the extent of introgression of the genome of the diverse parent in interespecific hybrids [40] or to evaluate progenies derived from interespecific crosses betweenT. aestivumandT. durum[41].
This paper evaluates the effectiveness of DArT as a high-throughput genotyping technology in tritordeum. This technology offers tritordeum breeding program an alternative approach to whole-genome profiling providing high quality markers that can be exploited in a range of molecular breeding and genomics applications in tritordeum. Additionally we set out to explore the possibility of using DArT array for genome background screening in tritordeum and to determine terms of discriminative ability using 1RS/1BL translocation lines.
Results
DArT array composition
A total of 2,372 out of 11,000 new DArT clones generated previously fromH. chilense[18] were printed in the array together with clones derived from other species developed in parallel projects (2,071 hexaploid wheat-derived markers, 290 fromH. vulgare, 208 from triticale and rye). From 4,941 clones identified on the array, a set of 3,357 markers were selected following the quality criteria explained above (P value and reproducibility), with 2,377 polymorphic markers used for diversity analyses of the 46 tritordeum lines (Additional file1).
Wheat-H. chilensechromosome addition lines allowed the physical location of a total of 2,209H. chilense-derived markers. Out of these, 1,280 markers could be assigned to a specificH. chilensechromosome on basis of two replicates (Additional file2).The remainder could not be assigned because a) they were also present in genomic representations generated from wheat; b) they were absent in representations generated from H1, the accession used to developH. chilense-wheat addition lines; c) they were found in H1 and absent in all the addition lines which suggests that these markers are located in either 2HchL or 3Hch, since addition lines carrying these chromosomes are not available or d) because they gave signal in two chromosomes and they were excluded of the study.
A subset of 450 DArT markers was shared with our mapping project [18], where Diversity Arrays technology (DArT) genomic libraries were developed fromH. chilenseaccessions. Out of these, 378 physically mapped to a specificH. chilensechromosome matched the quality criteria in the present work.
Genetic analysis
PIC ranged from 0.04 to 0.5, with an average of 0.31, for a bi-allelic marker, the minimum and maximum PIC values are 0 and 0.5, respectively. Two analyses were performed. The first of them included all the DArT markers while the second used polymorphic markers derived fromH. chilenseand D-genome, since we were looking for Hch/D chromosomes substitutions. The latter analyse produced a better discrimination among tritordeum lines and thus only results obtained using this subset of markers are shown. Brieftly, both the dendrogram and the PCoA analyses obtained with all markers included two non-substituted tritordeums with the substituted ones. Besides, the structure analysis separated between the substituted tritordeums and non-substituted ones. However, further analysis was required to differentiate among the different substitions and it was unable to distinguish the DS6D (6Hch) and other substitutions.
First, the 1,145 polymorphic markers derived fromH. chilenseand wheat D genome were considered to construct a dendrogram (Figure1).分为两个主要的cl登记入册usters with a cophenetic correlation value of 0.912 indicating an excellent fit of the similarity matrix data to the tree topology. The first group was formed by 27 accessions and it included those tritordeums with complete chromosome composition and the second group clustered 19 accessions carrying suspected or known chromosomal substitutions. Within this latter group, the inspection of the DArT markers dataset allowed to assign each subgroup to a different chromosome substitution. Five subgroups were differentiated, four of them comprised a single chromosome substitution (DS1D (1Hch), DS2D (2Hch), DS5D (5Hch) and DS6D (6Hch)) and one group had three genotypes with double chromosome substitution (DS2D (2Hch) and DS5D (5Hch)) (Figure1).
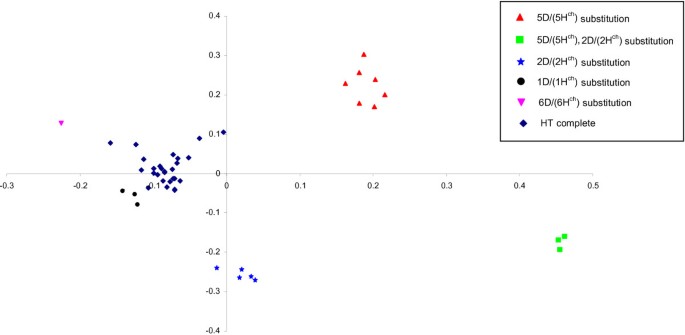
The principal coordinate (PCoA) was also constructed based on genotype data from the polymorphic DArT markers derived fromH. chilenseand wheat D genome. A two dimensional scatter plot of the 46 tritordeum genotypes, shown in Figure2, confirmed the five different subgroups found by UPGMA dendrogram although the DS6D (6Hch) and the DS1D (1Hch) substitution were placed near the non-substituted tritordeums. The two first dimensions of PCoA explained 34% and 25% respectively of the variation present in the genetic distance calculated between genotypes. Again, tritordeums carrying D/Hchsubstitutions were clearly differentiated from non-substituted tritordeums (Figure2).
Principal coordinate analysis of 46 tritordeum lines based on theH. chilenseand D-genome polymorphic DArT markers.The diagrams show the positions of individual accessions in the space spanned by the first two coordinates. The genetic composition of the lines is shown as follows: triangle: DS 5D (5Hch); square: double DS2D (2Hch) and 5D (5Hch); star: DS2D (2Hch); circle: DS1D (1Hch); inverted triangle: DS 6D (6Hch) and diamond: euploid tritordeum.
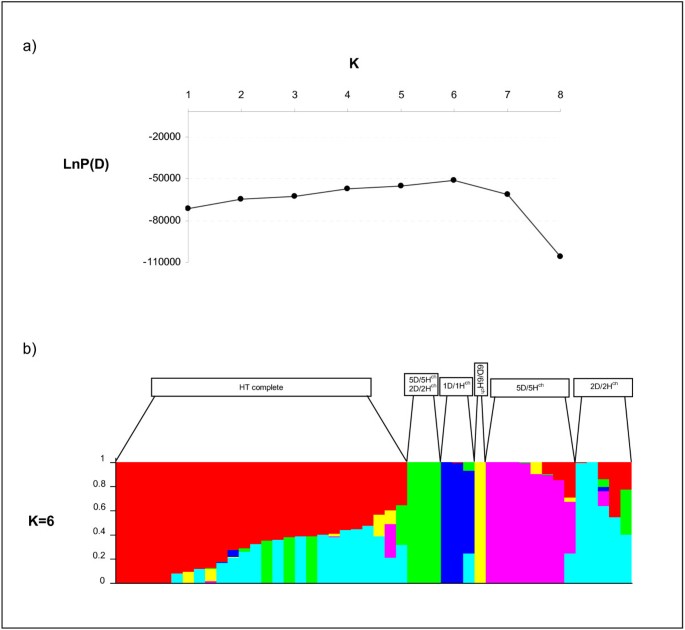
Genetic structure analysis detected an underlying structure, with six groups, based on the criterion of maximization of the natural log probability of the data, which is proportional to the posterior probability of K [42].The results pointed out to a clear cut-off point for the number of groups in our sample for K=6 (Figure3a). The genotypes were spread among the six groups as follows: Group I comprised 27 genotypes, including all tritordeums with complete chromosome composition, Group II includes three accessions from double chromosome substitution DS5D (5Hch) and DS2D (2Hch), Group III comprised three genotypes from DS1D (1Hch) chromosome substitution, Group IV included one line from DS6D (6Hch) substitution, Group V had seven genotypes from the DS5D (5Hch) chromosome substitution and group VI included five lines from the DS2D (2Hch) chromosome substitution (Figure3b).
Membership of tritordeum lines using DArT markers and the package STRUCTURE. a) Evolution of the natural log probability of the data, which is proportional to the posterior probability of K, against K (number of populations)b) Each individual is represented by a line partitioned in five coloured segments that represent the individual’s estimated membership fractions to each one of the six clusters.
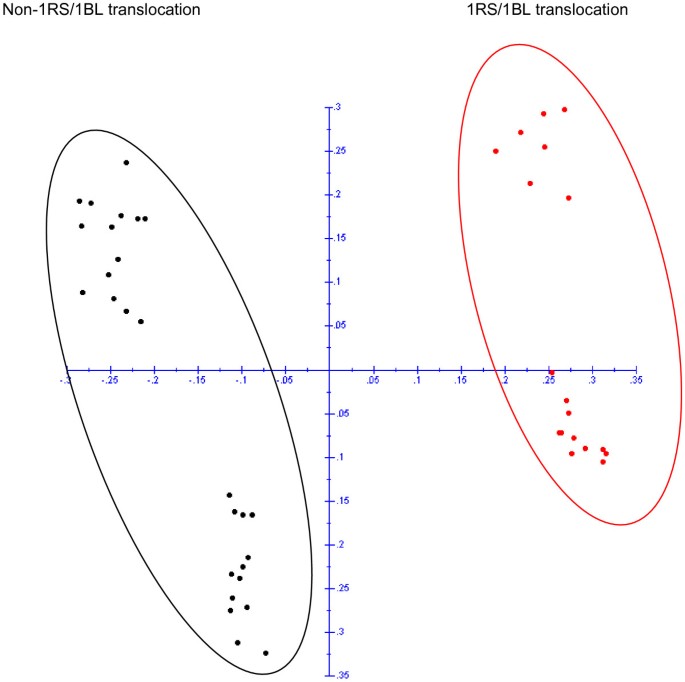
The potential of DArT markers to detect translocations was studied on the basis of 114 polymorphic DArTs markers derived from rye and Triticale. PCoA analysis of these markers which revealed a clear discrimination between 1RS/1BL and non-1RS/1BL translocation lines (Figure4).Once we confirmed the 1RS/1BL translocation with the specific primers for the locus SEC-1b, we inspected the DArT data file to identify markers discriminating the eighteen tritordeum lines carrying the 1RS/1BL translocation. Thirty three markers were only present in the tritordeums with the translocation 1RS/1BL (Additional file3).Most of them (31) were derived from rye or triticale and out of these; twenty eight were previously assigned to chromosome 1B and three not assigned to any chromosome. In addition, a single marker from each wheat (1B) and barley (1H) also discriminated between translocated and non-translocated lines. Similarly, chromosome specific EST and SSR specific markers (Table1) were used to verify the chromosome composition of the different lines studied. These markers either produce different amplification products inH. chilenseand wheat or exclusively amplify theH. chilensegenome, being therefore useful to detect chromosome substitutions.
Cytogenetic characterization by GISH
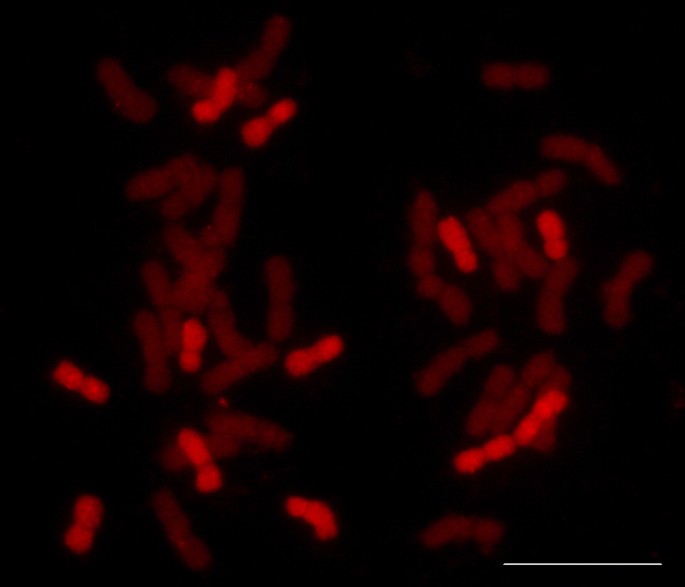
Genomic in situ hybridization was used to assess the chromosome substitutions usingH. chilensegenomic DNA as probe (detected with biotin). Sixteen single chromosome substitution lines were found to have only 12 chromosomes fromH. chilensecorresponding with DS1D (1Hch), DS2D (2Hch), DS5D (5Hch) and DS6D (6Hch) while three double chromosome substitution lines corresponding to DS2D (2Hch) and DS5D (5Hch) showed 10 chromosomes fromH. chilense(Figure5).The rest of the lines were unsubstituted tritordeums carrying all fourteenH. chilensechromosomes. In addition, eighteen lines showing 1RS/1BL translocations were detected usingS. cerealegenomic DNA as probe (data not shown).
Discussion
The availability of reliable molecular markers is of great importance for plant breeding. The ideal molecular marker technique should generate hundreds of molecular markers that cover the entire genome in a single, simple and reliable experiment [43].全基因组分子标记用于生殖plasm characterization, assessment of genetic diversity, to accelerate introgression or backcrossing programs, and for the mapping of complex traits. The high number of DArT markers generated in a single assay covering the whole genome allows the screening of a large number of genotypes in a rapid and efficient way.
Previous characterizations of the tritordeum have relied on cytogenetics using GISH and PCR-based markers [33,34].The GISH technique is laborious and unsuitable for high-throughput analyses. Similarly, the use of small sets of molecular markers may fail to detect translocations between Hchand D genomes in a high proportion and thus they only partially fulfil the requirements of our research programs. The development of DArT markers inH. chilensehas facilitated the use of these markers in tritordeum breeding program and related projects. In this work, we took advantage of the existence of a large number of DArT markers previously developed from hexaploid wheat,H. vulgare, Triticale,S. cerealeandH. chilense. A high percentage of DArT markers (48%) were polymorphic among the accessions studied which is higher than observed for DArT in other crops. For example a polymorphism rate of 9.4% observed in wheat [39飞镖的多态性标记并用时)ginating from hexaploid wheat, triticale, and rye as 8.6%, 23.4% and 23.8%, respectively in triticale [44].Since the DArT array was designed to maximize polymorphism rate considering previous experiments, this has resulted in a much higher polymorphism rate than reported in other crops. The average PIC value of 0.31 found in the current study was informative and comparable to values previously observed for barley (0.38) [38], wheat (0.31) [39], hop (0.34) [45], and triticale (0.37) [44].
H. chilenseand D-genome polymorphic markers were selected for genetic analysis to increase the power of detection of chromosome substitutions involving these genomes.
All three diversity analyses showed a good level of consistence to discriminate between substituted and non-substituted tritordeums. Both the dendrogram and the structure analysis grouped separately each type of chromosome substitutions but the first is much faster. Finally, the PCoA analysis discriminated well among groups. However, the DS6D (6Hch) and the DS1D (1Hch) groups appeared closely to non-substituted tritordeums which might lead to errors in classification in future analyses. In any case, we think that the application of DArT to the tritordeum breeding program would benefit from the analysis with all three methodologies since lines grouped differently in different methodologies may require a deeper analysis of the results.
The development of substitution lines has proven to be an efficient tool for the improvement of important agronomic traits. For instance, a major contribution to triticale improvement was achieved by the use of ‘Armadillo’, a line showing improved fertility due to the chromosome substitution DS2D (2R) [46].Although subsequent international yield trials indicated that complete hexaploid triticales showed a better agronomic performance than substitutes [47], the contribution of Armadillo to triticale improvement was very important. Similarly, D/(Hch)替换允许br的提高eadmaking quality [34] or free-threshing ability [33] but it is expected that tritordeums without chromosome substitutions will show superior performance over substituted ones in the future, as was observed in triticale. Nevertheless, the occurrence of translocations involving Hchand D chromosomes may still add interesting characteristics, as seen in wheat. Indeed, the 1RS/1BL translocation is one of the most frequently used alien introgresions in wheat-breeding programs throughout the world [48].The 1RS/1BL has positive effect on agronomic traits like yield performance, however, 1RS carries the Sec-1 locus coding for ϵ-secalin, which results in negative effects on bread-making quality. DArT markers clearly discriminated the lines with the 1RS/1BL translocation and thus, it is expectable that they will also allow the detection of D/(Hch) translocations when available. Accordingly these markers will prove very useful in parallel ongoing projects such as the development of hybrid wheat [49–51].
Conclusions
In conclusion, DArT markers allowed discrimination of the substitution lines involving the D genomes/Hch. All three methodologies clearly separated complete from substituted tritordeums but the combination between them will allow better discrimination of the specific lines. DArT markers also allowed the detection of translocations as evidenced from by the study of the 1RS/1BL translocation and, thus, they will very useful both in the tritordeum breeding program and the common wheat hybrid system.
Methods
Plant material
Forty-six accessions of hexaploid tritordeum originating from different stages of tritordeum breeding program were evaluated.Triticum aestivumcv. Chinese Spring (CS)-H. chilenseaddition lines for complete chromosomes 1Hch, 4Hch, 5Hch, 6Hchand 7Hch(named CS MA 1Hch-1HchS, CS DA4Hch, CS DA5Hch, CS DA6Hchand CS DA7Hch, respectively, where MA refers to monosomic addition and DA means disomic addition), and the wheat-(CS)-H. chilenseditelosomic addition lines CS DA1HchS, CS DA2HchS, CS DA5HchL, CS DA6HchS, CS DA7Hchα, CS DA7Hchβ were used to assign markers to specific chromosomes.T. aestivumcv ‘Chinese Spring’ andH. chilenseaccession H1 were also included. DNA was extracted from young leaf tissue from a single plant of each genotype using the protocol recommended by Triticarte Pty. Ltd. (http://www.triticarte.com.au).
Genotyping
A total of 4,941 DArT clones were printed in the array. Most of them were derived fromH. chilense(2,372) and hexaploid wheat (2,071). The array was completed with markers from barley (290) and rye and triticale (208). The resulting composite array was then used to fingerprint tritordeum and addition lines using the standard DArT protocol [52].Only DArT markers with a quality criteria, P value and reproducibility higher than 80 and 97% respectively, were selected.
Polymorphism information content (PIC), a measure of the informativeness of a genetic marker, was also calculated for each marker as follows; PIC = 1 − ΣPi2, where Pi is the frequency of theith allele in the examined genotypes. Chromosome substitutions in tritordeum were verified as previously described [33].A set of Expressed sequence tagged (EST) markers [23,24] and wheat chromosome-specific SSRs [53] were used to verify the presence ofH. chilenseand wheat chromosomes respectively (Table1).1RS/1BL translocations were detected using the primer pair SecA2/SecA3 designed specifically to amplify a sequence of ω-secalin gene (locus SEC-1b) located on the short arm of the rye chromosome 1R [54].
Data analysis
The data matrix containing the 0/1 scores of theH. chilenseand D-genome-derived polymorphic DArT markers was transformed to a genetic distance matrix using Jaccard’s coefficient [55].The genetic distance matrix was used to produce an unrooted Unweighted Pair Group Method with Algorithmic Mean (UPGMA) dendrogram using the program Dissimilarity Analysis and Representation for Windows, also known as DARwin [56].Also, the first two principal coordinates of the resulting Jaccard matrix were extracted to display the position of the accessions in a two-dimensional space (as an indication of the diversity of each pair of the accessions). Similarly, PCoA analysis was employed using polymorphic markers from rye and triticale genome.
Model-based clustering, employing a Bayesian algorithm, was applied to infer the genetic structure of the 46 tritordeum accessions using STRUCTURE software version 2.3.1 [57].The program was run assuming a population admixture model and correlated allele frequencies. The number of assumed groups (K) was set to vary between 1 and 10, and for each value of K five times independently MCMC (Markov Chain Monte Carlo) of 50,000 iterations was run in order to verify that the estimates were consistent across runs, of which the first 10,000 were discarded as burn-in. The likelihood of the data for a given number of assumed groups (K) is provided by the software, and the value of K with the highest likelihood can be interpreted to correspond to an estimate for the underlying number of groups. Within the admixture model we can obtain the membership probabilities of each genotype to each group inferred.
Genomic in situ hybridization (GISH)
Root tips of 1-cm length were collected from germinating seeds and pre-treated for 4h in an aqueous colchicine solution (0.05%) at 25°C. They were fixed in a freshly prepared 3 absolute ethanol: 1 glacial acetic acid (v/v) mixture and stored at 4°C during 1 month approximately. Preparations were made as described by [58].TotalH. chilensegenomic DNA was labelled by nick translation with biotin-11-dUTP (Roche Corporation, Basel, Switzerland) and totalSecale cerealegenomic DNA was labelled with digoxigenin-dUTP. Both probes were mixed in the hybridization solution to a final concentration of 5 ng/ml. Biotin- and digoxigenin-labelled probes were detected with streptavidin-Cy3 conjugates (Sigma, St Louis, MO, USA) and antidigoxigenin-FITC (Roche Corporate) respectively. Chromosomes were counterstained with DAPI (4′, 6-diamidino-2-phenylindole) and mounted in Vectashield (Vector Laboratories Inc.). Slides were examined by using a Zeiss LSM 5 Pa confocal laser scanning microscope with LSM 5 Pascal software version 3.0 (Zeiss, Jena, Germany).
References
- 1.
Friebe B, Jiang J, Raupp WJ, McIntosh RA, Gill BS: Characterization of wheat-alien translocations conferring resistance to diseases and pests: current status. Euphytica. 1996, 91: 59-87. 10.1007/BF00035277.
- 2.
Jiang J, Friebe B, Gill BS: Recent advances in alien gene transfer in wheat. Euphytica. 1994, 72: 199-212.
- 3.
Jones SS, Murray TD, Allan RE: Use of alien genes for the development of disease resistance in wheat. Annu Rev Phytopathol. 1995, 33: 429-443. 10.1146/annurev.py.33.090195.002241.
- 4.
Mares D, Mrva K: Genetic variation for quality traits in synthetic wheat germplasm. Aust J Agric Res. 2008, 59: 406-412. 10.1071/AR07224.
- 5.
Ogbonnaya FC, van Ginkel M, Brettell R: Preface: “Synthetics for Wheat Improvement”. Proceedings of the 1st Synthetic Wheat Symposium, September 2006. Aust J Agric Res. 2008, 59: 389-390. 10.1071/ARv59n5_PR.
- 6.
van Ginkel M, Ogbonnaya F: Novel genetic diversity from synthetic wheats in breeding cultivars for changing production conditions. Field Crops Res. 2007, 104: 86-94. 10.1016/j.fcr.2007.02.005.
- 7.
Rodríguez-Suárez C, Ramírez MC, Martín A, Atienza SG: Applicability of chromosome-specific SSR wheat markers for the introgression of Triticum urartu in durum wheat breeding programmes. Plant Genet Res Charact Utiliz. 2011, 9: 439-444. 10.1017/S147926211100061X.
- 8.
Kruse A: Hordeum x Triticum hybrids. Hereditas. 1973, 73: 157-161.
- 9.
Fedak G: Intergeneric hybrids with Hordeum. Barley: Genetics, Biochemistry, Molecular Biology and Biotechnology. Edited by: Shewry PR. Osford, UK: C.A.B. International, The Alden Press Ltd: 1992, 45-68.
- 10.
Kimber G, Sallee PJ: A trigeneric hybrid in the Triticeae. Cereal Res Commun. 1979, 7: 5-9.
- 11.
Martin A, Chapman V: A hybrid betweenHordeum chilenseandTriticum aestivum. Cereal Res Commun. 1977, 5: 365-368.
- 12.
Martín A, Sánchez-Monge Laguna E: Cytology and morphology of the amphiploidHordeum chilense’Triticum turgidumconvDurum. Euphytica. 1982, 31: 261-267. 10.1007/BF00028329.
- 13.
削R,雅各布森N,巴登C,约根森RB,林德e-Laursen I: An ecogeographical study of the genus Hordeum. Systematic and Ecogeographic Studies on Crop Genepools. 7. Rome, Italy: International Board of Plant Genetic Resources: 1995, 129.
- 14.
Rodríguez-Suárez C, Giménez MJ, Ramírez MC, Martín AC, Gutierrez N, Ávila CM, Martín A, Atienza SG: Exploitation of nuclear and cytoplasm variability inHordeum chilensefor wheat breeding. Plant Genet Res Charact Utiliz. 2011, 9: 313-316. 10.1017/S1479262111000402.
- 15.
Hernández P, Rubio MJ, Martín A: Development of RAPD markers in tritordeum and addition lines ofHordeum chilenseinTriticum aestivum. Plant Breed. 1996, 115: 52-56. 10.1111/j.1439-0523.1996.tb00870.x.
- 16.
Hernández P, Martín A, Dorado G: Development of SCARs by direct sequencing of RAPD products: a practical tool for the introgression and marker-assisted selection of wheat. Mol Breed. 1999, 5: 245-253. 10.1023/A:1009637928471.
- 17.
Atienza SG, Ávila CM, Martín A: The development of a PCR-based marker forPsy1fromHordeum chilense, a candidate gene for carotenoid content accumulation in tritordeum seeds. Aust J Agric Res. 2007, 58: 767-773. 10.1071/AR06338.
- 18.
Rodríguez-Suárez C, Giménez MJ, Gutiérrez N, Ávila CM, Machado A, Huttner E, Ramírez MC, Martín A, Castillo A, Kilian A, et al: Development of wild barley (Hordeum chilense)-derived DArT markers and their use into genetic and physical mapping. Theor Appl Genet. 2012, 124: 713-722. 10.1007/s00122-011-1741-2.
- 19.
Hernández P, Hemmat M, Weeden NF, Dorado G, Martín A: Development and characterization ofHordeum chilensechromosome-specific STS markers suitable for wheat introgression and marker-assisted selection. Theor Appl Genet. 1999, 98: 721-727. 10.1007/s001220051126.
- 20.
Castillo A, Budak H, Martín AC, Dorado G, Börner A, Röder M, Hernández P: Interspecies and intergenus transferability of barley and wheat D-genome microsatellite markers. Ann Appl Biol. 2010, 156: 347-356. 10.1111/j.1744-7348.2010.00391.x.
- 21.
Castillo A, Budak H, Varshney R, Dorado G, Graner A, Hernández P: Transferability and polymorphism of barley EST-SSR markers used for phylogenetic analysis inHordeum chilense. BMC Plant Biol. 2008, 8: 97. 10.1186/1471-2229-8-97.
- 22.
Hagras AAA, Kishii M, Sato K, Tanaka H, Tsujimoto H: Extended application of barley EST markers for the analysis of alien chromosomes added to wheat genetic background. Breed Sci. 2005, 55: 335-341. 10.1270/jsbbs.55.335.
- 23.
Hagras AAA, Kishii M, Tanaka H, Sato K, Tsujimoto H: Genomic differentiation of Hordeum chilense from H-vulgare as revealed by repetitive and EST sequences. Genes Genet Syst. 2005, 80: 147-159. 10.1266/ggs.80.147.
- 24.
Nasuda S, Kikkawa Y, Ashida T, Rafiqul Islam AKM, Sato K, Endo TR: Chromosomal assignment and deletion mapping of barley EST markers. Genes Genet Syst. 2005, 80: 357-366. 10.1266/ggs.80.357.
- 25.
Cabrera A, Martín A: Cytology and morphology of the ampliploidHordeum chilense(4x) xAegilops squarrosa(4x). Theor Appl Genet. 1991, 81: 758-760.
- 26.
Cherif-Mouaki S, Said M, Alvarez JB, Cabrera A: Sub-arm location of prolamin and EST-SSR loci on chromosome 1HchfromHordeum chilense. Euphytica. 2011, 178: 63-69. 10.1007/s10681-010-0268-y.
- 27.
Said M, Cabrera A: A physical map of chromosome 4H(ch) from H-chilense containing SSR, STS and EST-SSR molecular markers. Euphytica. 2009, 167: 253-259. 10.1007/s10681-009-9895-6.
- 28.
Martín A, Martínez C, Rubiales D, Ballesteros J:Tritordeum:triticale’snew brother cereal. Triticale: today and tomorrow. Edited by: Güedes-Pinto H, Darvey N, Carnide VP. Dordrecht, NL: Kluwer Academic Publishers: 1996,57-72.
- 29.
Martín A, Cubero JI: The use of Hordeum chilense in cereal breeding. Cereal Res Commun. 1981, 9: 317-323.
- 30.
Martín A, Alvarez JB, Martín LM, Barro F, Ballesteros J: The development of tritordeum: a novel cereal for food processing. J Cereal Sci. 1999, 30: 85-95. 10.1006/jcrs.1998.0235.
- 31.
Atienza SG, Ballesteros J, Martín A, Hornero-Mendez D: Genetic variability of carotenoid concentration and degree of esterification among tritordeum (xTritordeum Ascherson et Graebner) and durum wheat accessions. J Agri Food Chem. 2007, 55: 4244-4251. 10.1021/jf070342p.
- 32.
Ballesteros J, Ramírez MC, Martínez C, Atienza SG, Martín A: Registration of HT621, a high carotenoid content tritordeum germplasm line. Crop Sci. 2005, 45: 2662-2663. 10.2135/cropsci2004.0641.
- 33.
Atienza SG, Martín AC, Martín A: Introgression of wheat chromosome 2D or 5D into tritordeum leads to free-threshing habit. Genome. 2007, 50: 994-1000. 10.1139/G07-081.
- 34.
Ballesteros J, Ramírez MC, Martínez C, Barro F, Martín A: Bread-making quality in hexaploid tritordeum with substitutions involving chromosome 1D. Plant Breed. 2003, 122: 89-91. 10.1046/j.1439-0523.2003.00806.x.
- 35.
Cabrera A, Friebe B, Jiang J, Gill BS: Characterization ofHordeum chilensechromosomes by C-banding and in situ hybridization using highly repeated DNA probes. Genome. 1995, 38: 435-442. 10.1139/g95-057.
- 36.
Jaccoud D, Peng K, Feinstein D, Kilian A: Diversity arrays: a solid state technology for sequence information independent genotyping. Nucleic Acid Res. 2001, 29: e25. 10.1093/nar/29.4.e25.
- 37.
Kilian A, Huttner E, Wenzl P, Jaccoud D, Carling J, Caig V, Evers M, Heller-Uszynska CC, Patarapuwadol S, Xia L, et al: The fast and the cheap: SNP and DArT-based whole genome profilling for crop improvement. Proceedings of the international congress “In the wake of the double helix: from the green revolution to the gene revolution”, 27–31 May, 2003. Edited by: Tuberosa R, Philips RL, Gale M. Bologna, Italy: Avenue Media: 2005, 443-461.
- 38.
Wenzl P, Carling J, Kudrna D, Jaccoud D, Huttner E, Kleinhofs A, Kilian A, et al: Diversity Arrays Technology (DArT) for whole-genome profiling of barley. P Natl Acad Sci USA. 2004, 101: 9915-9920. 10.1073/pnas.0401076101.
- 39.
Akbari M, Wenzl P, Caig V, Carling J, Xia L, Yang S, Uszynski G, Moheler V, Lehmensiek A, Kuchel H, et al: Diversity arrays technology (DArT) for high-throughput profiling of the hexaploid wheat genome. Theor Appl Genet. 2006, 113: 1409-1420. 10.1007/s00122-006-0365-4.
- 40.
Kopecký D, Bartos J, Christelová P, Cernoch V, Kilian A, Dolezel J: Genomic constitution ofFestucaxLoliumhybrids revealed by the DArTFest array. Theor Appl Genet. 2011, 122: 355-363. 10.1007/s00122-010-1451-1.
- 41.
Eberhard FS, Zhang P, Lehmensiek A, Hare RA, Simpfendorfer S, Sutherland MW: Chromosome composition of an F2Triticum aestivumxT. turgidumspp.durumcross analysed by DArT markers and MCFISH. Crop Pasture Sci. 2010, 61: 619-624. 10.1071/CP10131.
- 42.
Falush D, Stephens M, Pritchard JK: Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003, 164: 1567-1587.
- 43.
Luikart G, England PR, Tallmon D, Jordan S, Taberlet P: The power and promise of population genomics: from genotyping to genome typing. Nature Reviews Genetics. 2003, 4: 981-994.
- 44.
Badea A, Eudes F, Salmon D, Tuvesson S, Vrolijk A, Larsson CT, Caig V, Huttner E, Kilian A, Laroche A: Development and assessment of DArT markers in triticale. Theor Appl Genet. 2011, 122: 1547-1560. 10.1007/s00122-011-1554-3.
- 45.
Howard EL, Whittock SP, Jakse J, Carling J, Matthews PD, Probasco G, Henning JA, Darby P, Cerenak A, Javornik B, et al: High-throughput genotyping of hop (Humulus lupulus L.) utilising diversity arrays technology (DArT). Theor Appl Genet. 2011, 122: 1265-1280. 10.1007/s00122-011-1529-4.
- 46.
Zillinsky FJ: The development of triticale. Adv Agron. 1974, 26: 315-348.
- 47.
Mergoum M, Pfeiffer WH, Peña RJ, Ammar K, Rajaram S: Triticale crop improvement: the CIMMYT programme. Triticale Improvement and Production Plant Protduction and Protection Paper 179. Edited by: Mergoum M, Gómez-Macpherson H. 2004, Rome: FAO, 11-26.
- 48.
Rabinovich SV: Importance of wheat-rye translocations for breeding modern cultivars ofTriticum aestivumL. (Reprinted from Wheat: Prospects for global improvement, 1998). Euphytica. 1998, 100: 323-340. 10.1023/A:1018361819215.
- 49.
Martín AC, Atienza SG, Ramírez M, Barro F, Martín A: Molecular and cytological characterization of an extra acrocentric chromosome that restores male fertility of wheat in the msH1 CMS system. Theor Appl Genet. 2010, 121: 1093-1101. 10.1007/s00122-010-1374-x.
- 50.
Martín AC, Atienza SG, Ramírez MC, Barro F, Martín A: Male fertility restoration of wheat inHordeum chilensecytoplasm is associated with 6H(ch)S chromosome addition. Aust J Agric Res. 2008, 59: 206-213. 10.1071/AR07239.
- 51.
Martín AC, Atienza SG, Ramírez MC, Barro F, Martín A: Chromosome engineering in wheat to restore male fertility in the msH1 CMS system. Mol Breed. 2009, 24: 397-408. 10.1007/s11032-009-9301-z.
- 52.
Kilian A, Wenzl P, Huttner E, Carling J, Xia L, Blois H, Caig V, Heller-Uszynska CC, Jaccoud D, Hopper C, et al: Diversity arrays technology: a generic genome profiling technology on open platforms. Methods Mol Biol. 2012, 888: 67-88. 10.1007/978-1-61779-870-2_5.
- 53.
Röder M, Korzun V, Wendehake K, Plaschke J, Tixier MH, Leroy P, Ganal MW: A microsatellite map of wheat. Genetics. 1998, 149: 2007-2023.
- 54.
de Froidmont D: A co-dominant marker for the 1BL/1RS wheat-rye translocation via multiplex PCR. J Cereal Sci. 1998, 27: 227-232.
- 55.
Jaccard P: Nouvelles recherches sur la distribution florale. Bull Soc Vand Sci Nat. 1908, 4: 223-270.
- 56.
Perrier X, Flori A, Bonnot F: Data analysis methods. Genetic diversity of cultivated tropical plants. Edited by: Hamon P, Seguin M, Perrier X, Glaszmann JC. Enfield: Science Publishers, Monpellier: 2003, 43-76.
- 57.
Pritchard JK, Stepehns M, Donnelly P: Inference of population structure using multilocus genotype data. Genetics. 2000, 155: 945-959.
- 58.
Prieto P, Ramírez MC, Ballesteros J, Cabrera A: Identification of intergenomic translocations involving wheat,Hordeum vulgareandHordeum chilensechromosomes by FISH. Hereditas. 2001, 135: 171-174.
Acknowledgments
This research was supported by grants AGL2009-11359 and 200840I137 from the Ministerio de Economía y Competitividad, CSIC and FEDER. This work is included within the CSIC – Agrasys SL collaboration agreement for the development of tritordeum.
Author information
Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AC performed SSR, EST and GISH genotyping, data analysis, interpretation of results and paper writing. MCR participated in chromosome counting, GISH genotyping and plant material development. ACM participated in SSR, EST and GISH genotyping. AK participated in the DArT genotyping. AM developed all the plant materials, conception, design of the study and interpretation of the results. SGA participated in design of the study, data analysis, interpretation of results and paper writing. All authors read and approved the final manuscript.
Electronic supplementary material
Polymorphic markers found among 46 tritordeum accessions.
Additional file 1:H. chilenseand D- genome markers used for genetic analysis are shown in bold. (XLSX 590 KB)
Markers assigned to a specific
Additional file 2:H. chilensechromosome using wheat-H. chilenseaddition lines.(XLSX 90 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open AccessThis article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Castillo, A., Ramírez, M.C., Martín, A.C.et al.High-throughput genotyping of wheat-barley amphiploids utilising diversity array technology (DArT).BMC Plant Biol13,87 (2013). https://doi.org/10.1186/1471-2229-13-87
Received:
Accepted:
Published:
Keywords
- Tritordeum
- DArT
- Chromosome substitution
- GISH
- EST
- SSR markers