- Research
- Open Access
- Published:
VRN1genes variability in tetraploid wheat species with a spring growth habit
BMC Plant Biologyvolume16, Article number:244(2016)
Abstract
Background
Vernalization genesVRN1play a major role in the transition from vegetative to reproductive growth in wheat. In di-, tetra- and hexaploid wheats the presence of a dominant allele of at least oneVRN1gene homologue (虚拟现实n-A1, 虚拟现实n-B1,虚拟现实n-G1or虚拟现实n-D1) determines the spring growth habit. Allelic variation between the虚拟现实n-1andvrn-1alleles relies on mutations in the promoter region or the first intron. The origin and variability of the dominantVRN1alleles, determining the spring growth habit in tetraploid wheat species have been poorly studied.
Results
Here we analyzed the growth habit of 228 tetraploid wheat species accessions and 25 % of them were spring type. We analyzed the promoter and first intron regions ofVRN1genes in 57 spring accessions of tetraploid wheats. The spring growth habit of most studied spring accessions was determined by previously identified dominant alleles ofVRN1genes. Genetic experiments proof the dominant inheritance of虚拟现实n-A1dallele which was widely distributed across the accessions ofTriticum dicoccoides. Two novel alleles were discovered and designated as虚拟现实n-A1b.7and虚拟现实n-B1dic. Vrn-A1b.7had deletions of 20 bp located 137 bp upstream of the start codon and mutations within the VRN-box when compared to the recessive allele ofvrn-A1. So far the虚拟现实n-A1dallele was identified only in spring accessions of theT. dicoccoidesandT. turgidumspecies.虚拟现实n-B1dicwas identified inT. dicoccoidesIG46225 and had 11 % sequence dissimilarity in comparison to the promoter ofvrn-B1. The presence of虚拟现实n-A1b.7and虚拟现实n-B1dicalleles is a predicted cause of the spring growth habit of studied accessions of tetraploid species. Three spring accessionst . aethiopicumK-19059,T. turanicumK-31693 andT. turgidumcv. Blancal possess recessive alleles of bothVRN-A1andVRN-B1genes.Further investigations are required to determine the source of spring growth habit of these accessions.
Conclusions
New allelic variants of theVRN-A1andVRN-B1genes were identified in spring accessions of tetraploid wheats. The origin and evolution ofVRN-A1alleles in di- and tetraploid wheat species was discussed.
Background
Flowering time is a critical agronomical trait that has a major impact on the adaptation to local climate and environmental conditions and grain yield in wheat species. Wheat cultivars differ in their requirements for extended exposure to low-temperature (vernalization) to initiate the transition from vegetative growth to flowering [1]。Ancestors of wheat, as well as modern wheat species with a winter growth habit, are planted in autumn and flower during the subsequent spring. These species require vernalization for transition from vegetative to reproductive growth. The vernalization requirement prevents the fragile flower meristems from being damaged by low temperatures and ensures that flowering occurs under optimal conditions in spring. Modern wheat cultivars with spring growth habit lack this vernalization requirement and can be planted in spring [2]。
Vernalization-induced flowering in wheat is mainly controlled by the vernalization genesVRN1,VRN2,VRN3andVRN4which interact with each other as well as other flowering control pathways [1–8]。VRN1genes mapped to the long arms of the 5 homoeological group chromosomes play a central role in complex vernalization pathways [9–12]。The floral activatorVRN1encodes a MADS-box transcription factor that is required for the initiation of reproductive development at the shoot apical meristem [3,13,14]。The expression ofVRN1occurs at a low basal level but a measurable increase is seen during prolonged treatment with low temperatures [3,13,14]。
In di-, tetra- and hexaploid wheats the presence of a dominant allele of at least oneVRN1gene homologue (虚拟现实n-A1,虚拟现实n-B1,虚拟现实n-G1or虚拟现实n-D1) determines the spring growth habit. Allelic variation between the虚拟现实n-1andvrn-1alleles relies on mutations in the promoter region or the first intron.
Analysis of diploid wheat species revealed three recessive allelesvrn-Am1,vrn-A1uandvrn-Am1bwhich determine the winter growth habit [3,15–18]。Thevrn-Am1allele is distributed in all diploid wheat species and represents the only allelic variant so far identified inTriticum sinskajaeA. Filat. et Kurk. [3,15–18]。Thevrn-A1uofT. urartuThum. ex Gandil. is identical tovrn-A1of winter accessions of polyploid wheats and differs fromvrn-Am1by a short deletion in the promoter [16,17]。Thevrn-Am1bwith a 48-bp deletion in theVRN1promoter in compare tovrn-Am1so far was found only in accessions ofT. monococcumL. [3,18]。In diploid wheatT. monococcumseveral dominant虚拟现实n-A1alleles which possess variable mutations in the promoter and/or first intron region were identified [3,15–18]。Two dominant alleles from diploid wild wheatT. boeoticumBoiss.虚拟现实n-Am1fand虚拟现实n-Am1a(虚拟现实n-A1h) posses short deletions in the promoter region [16,17]。No dominant alleles were identified inT. urartu[16,17]。
In tetraploid wheat species of sectionsTimopheeviiA. Filat. et Dorof. andDicoccoidesFlaksb. five recessive alleles ofVRN1genes (vrn-A1(vrn-A1u),vrn-A1b.3,vrn-A1b.4,vrn-B1andvrn-G1) were identified [16,17,19]。The sequences ofvrn-B1andvrn-G1alleles were identical to each other [16]。年代everal dominant and recessive alleles ofVRN-A1,VRN-B1andVRN-G1genes were identified in tetraploid wheat of two sectionsTimopheeviiA. Filat. et Dorof. andDicoccoidesFlaksb. [16,17,19–22]。Thus, analysis of tetraploid wheatsT. timopheevii(Zhuk.) Zhuk. andT. araraticumJakubz. revealed one dominant allele of theVRN-A1gene (虚拟现实n-A1f) comprising two deletions in the promoter region, and the only dominant allele ofVRN-G1gene (虚拟现实n-G1a) with the insertion in the promoter region [16]。Ten various dominant alleles of theVRN-A1gene which possess different mutations in compare tovrn-A1allele were identified in different tetraploid wheat species of sectionDicoccoidesFlaksb. Seven of them possess deletions of variable length in the promoter (虚拟现实n-A1b(虚拟现实n-A1b.1),虚拟现实n-A1b.2,Vrn-A1b.5,虚拟现实n-A1b.6, Vrn-A1e,虚拟现实n-A1fand虚拟现实n-A1d), one (虚拟现实n-A1i)有核苷酸替换和一个(虚拟现实n-A1a(虚拟现实n-A1a.3)) has a foldback element insertion in the promoter region [16,17,19,20]。The first intron sequence of theVRN-A1gene fromT. durumDesf. cultivar Lebsock was found to contain a large deletion identical toT. durum‘Langdon’ [21,22]。Further analysis allowed to identify this allele in accessions ofT. turgidumL.,T. carthlicumNevski,T. polonicumL., T.dicoccoides(科恩。亚瑟。et Graebn)。年代chweinf. andT. durumDesf. [17,19]。Novel allelic variantVRN-A1f-like identified by Ivaničová et al. [23] possess mutations in both promoter and first intron regions ofVRN-A1, while only mutations within first intron is the reason of the spring growth habit ofT. militinaeaccession.
In tetraploid wheats of the sectionDicoccoidesfour dominant alleles ofVRN-B1for which the variability is characterized by the mutations in the promoter region (insertion of repeated elements or short deletions) [16,19,21]。The虚拟现实n-B1ais the only dominant allele with the large deletion in the first intron which was identified in theDicoccoidesaccessions [19,22]。The only dominant allele虚拟现实n-G1afrom sectionTimopheeviiis characterized by foldback element insertion in the promoter region [16]。
For a long timevrn-A1was the only recessive allele identified in hexaploid wheat, but recently additional allelevrn-A1b.3was identified inT. vavilovii(Thum.) Jakubz. andT. speltaL [19]。年代even dominant alleles of theVRN-A1gene,虚拟现实n-A1a.1,虚拟现实n-A1a.2,虚拟现实n-A1b (Vrn-A1b.1),虚拟现实n-A1b.2,虚拟现实n-A1b.6,虚拟现实n-A1cand虚拟现实n-A1fwere found in hexaploid wheat [16,19,20,22,23]。The majority of the spring cultivars carry a虚拟现实n-A1a.1allele that has a miniature inverted-repeat transposable element (MITE) insertion and duplication in the promoter region [19]。虚拟现实n-A1ballelic variants and虚拟现实n-A1fallele have mutations and deletions of variable lengths in the promoter region, whereas虚拟现实n-A1chas a deletion in the first intron in comparison to the recessivevrn-A1allele [16,19,20,22,23]。Most of the dominant alleles ofVRN-B1andVRN-D1genes possess deletions in the first intron (虚拟现实n-B1a,虚拟现实n-B1b,虚拟现实n-B1cand虚拟现实n-D1a) [16,19,22,24–27]。年代everal recently identified alleles (虚拟现实n-B1ins, Vrn-D1c, Vrn-D1s) are characterized by different insertions within the promoter region [19,28,29]。The虚拟现实n-D1bis characterized by the deletion in intron 1 identical to虚拟现实n-D1aallele and a single nucleotide mutation at promoter and is associated with facultative growth habit [30]。
The evolution of spring cultivars of wheats from winter ancestors is a key event in the post-domestication spread of wheat [1]。However, studies of the major vernalization geneVRN1are mostly limited to the analysis of di- and hexaploid wheat species. In the present study we investigate the growth habit and variability of promoter and first intron regions ofVRN1genes in accessions of twelve tetraploid wheat species of sectionsDicoccoidesandTimopheevii.
Methods
Plant material
Accessions of 12 tetraploid wheat species were obtained from the following gene banks: N.I. Vavilov Institute of Plant Genetic Resources (VIR, Russian Federation), The Federal Research Center Institute of Cytology and Genetics SB RAS (Russian Federation), the National Small Grains Collection (NSGC, USA), International Center for Agricultural Research in the Dry Area (ICARDA, Syria), Kyoto University (Japan). Place of origin, specimen voucher and growth habit of each accession are presented in Additional file1: Table S1.
Greenhouse experiments
The growth habit of tetraploid wheat species was evaluated by growing in the greenhouse at 20–25 °C under a long photoperiod (18 h light) without vernalization treatment. F1hybrids of tetraploid near-isogenic lineT. dicoccumBlack SpringVRN-B1Emmer (i: BS2E) andT. dicoccum(Schrank) Schuebl. cv. Black Winter Emmer (BWE) were used as controls. In i: BS2E, spring growth habit is determined by the dominant allele of the虚拟现实n-B1gene [31]。T. dicoccumcv. BWE has a winter growth habit. F1hybrids of NIL BS2E withT. dicoccumcv. BWE have the genotype虚拟现实n-B1/vrn-B1and represent the latest maturing spring form at the border between spring vs. winter phenotypes, according to Pugsley [32] and Goncharov [33]。Accessions that headed before F1hybrids were classified as spring, whereas accessions that remained in the vegetative phase were classified as winter. A detailed procedure is described in Goncharov [33]。
To determine days to heading of spring accessions ten plants of each accession were grown in the greenhouse at 20–25 °C under a long photoperiod (18 h light) without vernalization treatment. Mean number of days to heading (χ ± s) and mean error (s) were estimated using standard Microsoft office software.
The number of dominantVRNgenes in tetraploid wheat was identified based on the segregations in the F2generations. F1hybrids between six accessions of tetraploid wheats and three tester lines, winter accessionsT. dicoccumcv. BWE,T. dicoccumBlack SpringVRN-A1Emmer (i: BS1E) and i: BS2E, were produced by emasculation of mother’s plant spikes and pollination with flowering father plant spikes using twirl-method.T. dicoccumcv. BWE has recessivevrn-A1andvrn-B1alleles while two near-isogenic test lines are characterized with the only specific dominant allele:虚拟现实n-A1in i: BS1E and虚拟现实n-B1of i: BS2E [32]。The segregation into spring versus winter forms for each cross was identified and compared with the expected segregation ratio using the Pierson chi-square test.
隔离总DNA, PCR扩增,克罗ning and sequencing
Total DNA was isolated from 100 mg of leaves using the DNeasy Plant Mini Kit (QIAGEN) according to the manufacturer’s protocol. A set of primers were used to amplify the promoter and first intron sequences ofVRN-A1,VRN-B1andVRN-G1genes (Additional file1:表S2)。
Polymerase chain reactions (PCR) were performed in a 20 μl volume with 10 mM Tris–HCl (pH 8.9), 1 mM (NH4)2年代O4, 4 mM MgCl2, 200 μM of each dNTP, 0.5 μM of each primer, 1 unit of Taq DNA polymerase and 0.1 μg of genomic DNA. The PCR program included an initial denaturation step for 3 min at 94 °C and 33 cycles of amplification consisting of 30 s denaturation at 94 °C, 40 s annealing at 52 °C, and 1 min extension at 72 °C. PCR products were separated by agarose gel electrophoresis and purified using a QIAquick Gel Extraction Kit (QIAGEN). Purified fragments were cloned into a pGEM®-T Easy vector using a pGEM-T Easy kit (Promega) and amplified with M13 primers (M13F = 5′-GTTTTCCCAGTCACGAC-3′, M13R = 5′-AGCGGATAACAATTTCACACAGGA-3′). Sequencing reactions were performed with 200 ng of the product and ABI BigDye Terminator Kit on an ABI 3130XL Genetic Analyser (Applied Biosystems) in SB RAS Genomics Core Facility (http://www.niboch.nsc.ru/doku.php/corefacility). In total 10 clones were sequenced for each target region of all wheat accessions with spring growth habit.
The sequences of the promoter and first intron region ofVRN-A1,VRN-B1andVRN-G1genes were deposited in GenBank (accession numbers are given in Tables1and2).
年代equence analyses
Nucleotide sequence alignments were performed using Vector NTI AdvanceTM version 10.0 program and improved with the MUSCLE algorithm in UGENE software (http://ugene.unipro.ru/) [34,35]。
Results
Growth habit of tetraploid wheat species
To analyze the variability of the promoter and first intron regions ofVRN-A1,VRN-B1andVRN-G1genes we chose a number of tetraploid wheat accessions, covering all species from bothTimopheeviiandDicoccoidessections (Additional file1: Table S1).
The growth habit of 228 accessions of tetraploid wheat species was checked by comparison to F1hybrids of i: BS2E and winterT. dicoccumcv. BWE (see Methods section). In total, 57 accessions of 10 tetraploid wheat species headed before the hybrids and thus revealed a spring growth habit (Table1; Additional file1: Table S1). For each of 57 spring accessions 10 plants were grown under glasshouse conditions and days to heading were recorded (Table1). 171 accessions ofT. dicoccoides(科恩。亚瑟。et Graebn)。Schweinf。T. ispahanicumHeslot andT. karamyscheviiNevski did not produce shoots without vernalization, thus confirming a winter growth habit (Additional file1: Table S1).
VRN-A1promoter region variability
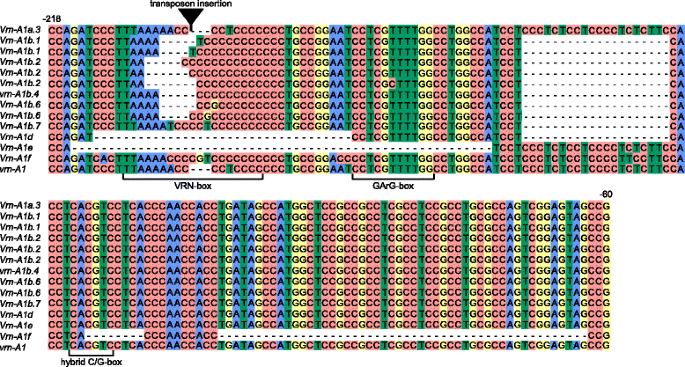
Mutations within the promoter region appear to be responsible for the major differences between dominant and recessive alleles of theVRN1gene and the cause of spring growth habit in wheat plants. Here, 57 accessions were screened using PCR amplification with A genome-specific primers designed by Yan et al. [20]。The PCR product of estimated length (~700 bp) was obtained for all species studied. After cloning of PCR products, all clones obtained were sequenced and analyzed. In total, 69 sequences of theVRN-A1promoter region were identified. New sequences were aligned together with known recessive and dominant allelic variants of theVRN-A1promoter region in di-, tetra- and hexaploid wheat obtained from GenBank. Comparative analyses demonstrated that the most variable zone of theVRN-A1promoter is located in the region from –63 to –220 bp (Fig.1).
Alignment ofVRN-A1gene promoter sequences identified from tetraploid wheats in the study. Transposon insertion is indicated by black triangle. Numbers of the nucleotides upstream from the start codon are given in accordance with the sequencevrn-A1(GenBank Ac.No GQ451819). Predicted regulatory regions are marked according to Golovnina et al. [16] and Muterko et al. [19]
Nine known allelic variants of theVRN-A1promoter虚拟现实n-A1a.3,虚拟现实n-A1b,虚拟现实n-A1b.2,虚拟现实n-A1b.6,虚拟现实n-A1d,虚拟现实n-A1e,虚拟现实n-A1f,vrn-A1b.4, andvrn-A1were identified. Promoter sequences ofVRN-A1from 17 accessions oft . aethiopicumJakubz.,T. dicoccoides,T. dicoccum,T. durumDesf. andT. turgidumL. were identical to the recessive allelevrn-A1identified in polyploids andvrn-A1uofT. urartu(Table1, Fig.1). Four accessionst . aethiopicumK-19059,T. turanicumJakubz. K-31693,T. turgidumcv. Blancal (K-20416) andT. turgidumK-11597 contained one nucleotide substitution in the promoter when compared to thevrn-A1sequence (GQ451819).
The other 36 accessions had mutations in the promoter region distinguishing them from the recessive allelic variantvrn-A1. The promoter region of theVRN-A1gene inT. dicoccoidesK-15900 and i: BS1E was identical to the虚拟现实n-A1a.3allele, and had an insertion of 231 bp in length (Table1, Fig.1). Five variants of虚拟现实n-A1ballele were identified in accessionsT. dicoccumcv. Dichter Rotlicher (K-1730),T. durumK-13768,T. turgidumk - 3047和k - 13489,T. polonicumK-19597 and K-43335. All sequences of虚拟现实n-A1bpromoter had common mutation which distinguish them fromvrn-A1sequence but vary in the length and sequences of VRN-box (Table1, Fig.1). Sequences ofT. durumK-13768 andT. dicoccumcv. Dichter Rotlicher (K-1730) were identical to previously described variants虚拟现实n-A1b.1and虚拟现实n-A1b.2, correspondingly.VNR-A1promoter sequences ofT. turgidumK-13489 and K-3047,T. polonicumK-19597 and K-43335 were identical to each other and虚拟现实n-A1b.6等位基因中确定四倍体和六倍体wheat species [19]。
Five accessions ofDicoccoidessection had two variants of the虚拟现实n-A1promoter sequence, both of which corresponded to the variants of虚拟现实n-A1ballele (Table1). Three accessionsT. durumK-18118 and IG 85879, andT. polonicumL. K-17893 possess identical set of promoter sequences:虚拟现实n-A1b.1variant identified previously and sequence differed from虚拟现实n-A1b.1only by insertion of one nucleotide “C” in C-rich segment of VRN-box (Fig.1). Two sequences from accessions ofT. dicoccoidesIG346783 andT. dicoccumK-7500 represent虚拟现实n-A1b.2variant but vary in the length of C-rich segment (Fig.1).
TheT. timopheeviiKU107-1, 38555 k - 29540 k - 8和50个基点删除ions as in the虚拟现实n-A1fsequence reported inT. araraticum(GQ451762) [16] (Table1, Fig.1). One accession ofT. araraticumK-30234 contained two sequences of the虚拟现实n-A1fallele which varied in the length of the C-rich segment of VRN-box.
We observed a 54 bp deletion in the promoter region ofT. carthlicumK-7106 that was identical to the虚拟现实n-A1ealleles ofT. durum(GQ451821) andT. dicoccum(AY616463) (Table1, Fig.1).
Eleven accessions ofT. dicoccoideshad two different deletions one 20 bp in length between -136 and -157 and another one 32 bp in length between -179 and -212 nucleotides upstream of the start codon and were identical to the虚拟现实n-A1dallele [17,20] (Table1, Fig.1).
The most interesting were two groups of accessions which possessed two different variants of虚拟现实n-A1promoter sequences. First group of accession includeT. dicoccoidesPI428105, PI352324 and PI352328 andT. araraticumK-58667 which are characterize by the presence of虚拟现实n-A1freported inT. araraticum(GQ451762) and one of two variants虚拟现实n-A1ballele (Table1). Second promoter sequences identified inT. dicoccoidesPI428105 and PI352328 were identical to recessive variantvrn-A1b.4. In case of theT. dicoccoidesPI352324 andT. araraticumK-58667 s sequences differed fromvrn-A1by 20 bp deletion located 137 bp upstream of the start codon, “A- > T” replacement and “CCC” insertion within the VRN-box. This variant was designated as虚拟现实n-A1b.7(Fig.1). Second group of accessions includeT. dicoccoidesPI467027, PI467014 and PI467019 possess two sequences ofVRN-A1promoter:虚拟现实n-A1dand虚拟现实n-A1falleles. Schematic representation of allVRN1promoter sequences identified in di- and tetraploid species is presented in Additional file2: Figure S1. Geographical distribution of theVRN-A1alleles identified in the studied samples is presented on Additional file2: Figure S2.
VRN-A1first intron variability
First intronVRN-A1gene in tetraploid wheats was analyzed using three primer pairs (Additional file1:表S2)。The first primer pair (Ex1/C/F and Intr1/A/R3) allowed us to amplify the sequences of theVRN-A1first intron which possesses a deletion of a characteristic length previously described forT. durum‘Langdon’ [22]。The short PCR product (~480 bp in length) containing the deletion was obtained for 7 accessions oft . aethiopicum(K-19301, K-19398, K-19553, K-19253, K-19650, K-43766 and St56), 1 accession ofT. dicoccoides(K-62328), 3 accessions ofT. dicoccum(K-20749, K-1730 and K-40306), 4 accessions ofT. durum(K-17784, K-17787, K-52989 and cv. Langdon), 1 accession ofT. polonicum(K-17893) and 2 accessions ofT. turgidum(cv. Maiorka (K-16156) and cv. Zafrani (K-11597)) (Table1). These sequences were identical to the虚拟现实n-A1callele ofT. turgidum‘Langdon’, possessing a 7222-bp deletion from 391 bp to 7612 bp compared to theT. aestivumNIL Triple Dirk Cvrn-A1allele (AY747600) [22]。
No positive results of PCR amplification were obtained with the primers Intr1/A/F2 and Intr1/A/R3. Thus, no sequences of theVRN-A1first intron containing the deletion identified in hexaploid Afghanistan landrace IL369 were presented among analyzed species. The last primer pair (Intr1/C/F and Intr1/AB/R) allowed us to amplify the intact sequences of the first intron, producing a product of the expected length (~1000 bp) for 39 tetraploid wheat accessions (Table1).
VRN-B1andVRN-G1promoter region variability
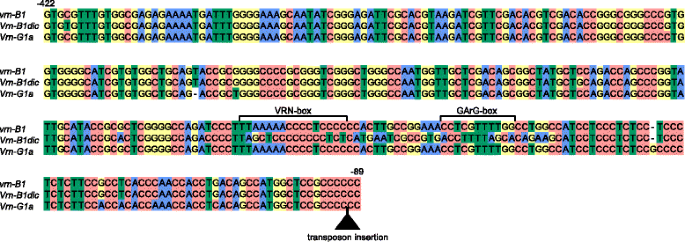
The B genome-specific primers were used to amplify and sequence the promoter region of five accessions of sectionDicoccoidesspecies,with recessivevrn-A1promoter sequences and an intactVRN-A1first intron. The fragment of the expected length ~ 1200 bp was obtained for all analyzed species. Further comparative analyses demonstrated that almost all clones from accessions ofDicoccoidessection contained sequences of the intactvrn-B1allele (Table2). The only exception wasT. dicoccoidesIG46225, for which two different clones were identified. The first clone corresponded to the recessivevrn-B1alleles identified previously [16,21,22]。The second clone differed from the recessive allele by 29 nucleotide substitutions, one deletion and one insertion of a single nucleotide in the region from -220 to -155 bp upstream from the start codon (11 % of dissimilarity) (Table2, Fig.2, Additional file2: Figure S1). The present allelic variant was different from all known B genome alleles ofVRN1gene and was named虚拟现实n-B1dic. NoVRN-B1promoter sequences containing retrotransposon insertions, previously described by Chu et al. [21], were identified among analyzed accessions.
Alignment ofVRN-B1andVRN-G1genes promoters identified from tetraploid wheats in the study. Transposon insertion is indicated by black triangle. Numbers of the nucleotides upstream from the start codon are given in accordance with the sequencevrn-B1(GenBank Ac.No AY616453). Predicted regulatory regions are marked according to Golovnina et al. [16] and Muterko et al. [19]
TheVRN-G1promoter was analyzed for five accessions of sectionTimopheeviispecies (Table2). Sequences of theVRN-G1promoter were identified forT. araraticum(accession K-30234) andT. timopheevii(accessions K-38555, K-29540 and K-29540). The 3′ part of these sequences was identical to the sequence of虚拟现实n-G1aallele fromT. timopheeviiK-38555 (GQ451755), while the 5′ end was identified for the first time and differed fromvrn-G1promoter by two deletions: 8 and 74 bp in length (Fig.2; Additional file2: Figure S1). The recessivevrn-G1promoter sequences were identified only for accession K-58667 ofT. araraticum.
VRN-B1andVRN-G1first intron variability
First introns ofVRN-B1andVRN-G1genes were analyzed using two primer pairs (see the Methods section). Sequences of theVRN-B1first intron possessing the large deletion described forT. aestivumNIL Triple Dirk B [22] were amplified with the primers Intr1/B/F and Intr1/B/R3. Positive results of the PCR amplification with this primer pair were obtained for one out of ten studied accessions (Table2). The sequence oft . aethiopicumK-18999 differs from the first intron sequence ofT. aestivumNIL Triple Dirk B (AY747603) by two nucleotide substitutions. The remaining 9 accessions gave positive results with the second primer pair (Intr1/B/F and Intr1/B/R4), and intact sequences ofVRN-B1andVRN-G1first introns (~1150 bp in length) were amplified (Table2).
Genetic control of growth habit in tetraploid species ofDicoccoidessection
Monogenic or digenic control of growth habit in some spring accessions of tetraploid wheat species ofDicoccoidessection was analyzed in the genetic experiments. Accessions of tetraploid species were crossed withT. dicoccumcv. Black Winter Emmer (BWE) which has recessivevrn-A1andvrn-B1alleles and two near-isogenic tester lines, which are characterized with specific dominant虚拟现实n-A1(i: BS1E) or虚拟现实n-B1(i: BS2E) alleles. It was demonstrated that spring growth habit of six accessions ofT. dicoccoides,T. dicoccumandT. durumis controlled by a single dominant geneVRN-A1(Table3). F2hybrids ofT. dicoccumcv. Dichter Rotlicher (K-1730),T. dicoccumcv. Bastard Emmer verastelter (K-40306) andT. durumcv. Langdon, K-17784 and K-17787 with BWE showed the segregation ration 3 to 1, while their F2hybrids with BS1E showed no segregation and spring growth habit (Table3). Thus, the results confirmed that the虚拟现实n-A1callele is dominant. ForT. dicoccoidesICG №23 monogenic control of spring growth habit was shown (Table3). All F2hybrids ofT. dicoccoidesICG №23 with BS1E showed spring growth habit which allows us to confirm that newly identified虚拟现实n-A1dallele is dominant.
A correlation between certain dominant variant ofVRN-A1genes and number of days to heading as well as a correlation between certain species and number of days to heading were not identified for the studied accessions of tetraploid wheats (Additional file2: Figure S3, Figure S4).
Discussion
Predicted source of the spring growth habit among tetraploid wheat species
Variability in the growth habit (spring vs. winter) of tetraploid wheat has been studied in greenhouse tests and 57 spring accessions were subsequently identified. For four of the species analyzed (T. polonicum,T. carthlicum,T. aephiopicum, andT. timopheevii) no winter accessions were identified in the present study nor in a previous study by Goncharov [36]。All studied accessions ofT. ispahanicumHeslot andT. karamyscheviiNevski were found to have a winter growth habit. The following analysis of theVRN-A1, VRN-B1andVRN-G1genes in accessions with a spring growth habit revealed the presence of different mutations within the promoter or first intron region of those genes.
For 36 of the spring accessions studied, we identified variability within the promoter region of theVRN-A1gene (Table1).VRN-A1promoter sequences of these 36 accessions matched one of four different dominant alleles (虚拟现实n-A1a.3,虚拟现实n-A1d,虚拟现实n-A1e,虚拟现实n-A1f) or one of four虚拟现实n-A1ballele variant (虚拟现实n-A1b.1, Vrn-A1b.2, Vrn-A1b.6,虚拟现实n-A1b.7). The presence of虚拟现实n-A1a,虚拟现实n-A1b.1, Vrn-A1b.2, Vrn-A1b.6,虚拟现实n-A1d,虚拟现实n-A1eor虚拟现实n-A1fwas previously predicted to be a determinant of the spring growth habit in wheat species [16,17,19,20]。
年代even of the 36 spring accessions studied possess two different allelic variants ofVRN-A1, at least one of which was dominant and could led to the spring growth habit. The presence of two different alleles in one accession could be explained by heterozygosity of the plant material or the variation of copy number of genes due to the duplication of the investigated region or the part of the genome. Presence of two different alleles has not been described for diploid or hexaploid wheat species, but this has been identified in the wild tetraploid species [16,37]。
Allelevrn-A1and variants of虚拟现实n-A1bwere the most frequently occurring, and were identified for 21 and 15 spring tetraploid accessions, respectively (Table1).虚拟现实n-A1dand虚拟现实n-A1fwere also common and presented in 14 and 11 spring tetraploid accessions, respectively. Dominant inheritance of虚拟现实n-A1dwas confirmed in the genetic experiments. The remaining two alleles are rare, the虚拟现实n-A1awas identified in two accessions while虚拟现实n-A1ewas found only once (Table1).
21 accessions oft . aethiopicum,T. dicoccoides,T. dicoccum,T. durum,T. polonicum,T. turanicumandT. turgidumcontained the recessive allele of theVRN-A1promoter. Therefore, its spring growth habit could be explained by other changes in theVRN1gene sequences, this may include mutations in theVRN-B1promoter regions, as well as in the first intron sequence of bothVRN-A1andVRN-B1genes. 16 out of 21 accessions showed the presence of a large deletion within theVRN-A1first intron region (虚拟现实n-A1c) and dominant inheritance of this allele was confirmed in the genetic experiments (Table1, Table3). Only one of the studied accessions possessed a disruption within theVRN-B1promoter sequence.T. dicoccoidesIG46225 contains two differentVRN-B1promoters: the first one was identical to the intact sequence of thevrn-B1allele, whereas the second one displayed a new allelic variant, named虚拟现实n-B1dic. The presence of the虚拟现实n-B1dicallele could be the cause of spring growth habit inT. dicoccoidesIG46225.虚拟现实n-B1dicallele is characterized by unexpected high dissimilarity in compare to thevrn-B1allele. If we exclude the deletions and insertions cases the other dominant alleles ofVNR1genes of di- and polyploidy wheats differ from recessive alleles by several SNPs [19,22,23,27,29]。另外,we could suggest that the虚拟现实n-B1dicrepresent the pseudogene copy originated by duplication within one genome. Previously, the investigation of the bread wheat genome showed the major impact of single gene duplications on the wheat evolution [38]。Moreover the gene duplications followed by gene loss, subfunctionalization or neofunctionalization played significant role in the evolution of MADS-box transcription factors [39]。Investigation of the虚拟现实n-B1dicexpression is required to proof the hypothesis.
t . aethiopicumK-18999 was the only accession for which we identified intact promoters ofVRN-A1andVRN-B1genes, and an intact first intron of theVRN-A1gene. A deletion has been found in the first intron of theVRN-B1gene. This deletion is a predicted cause oft . aethiopicumK-18999’s spring growth habit.
One group of accessions is of particular interest in the investigation of the possible cause of spring growth habit in tetraploid wheat species.T. dicoccumcv. Dichter Rotlicher (K-1730) andT. polonicumK-17893, both of which posses the dominant allele of theVRN-A1promoter, but contain deletions in the first introns of theVRN-A1gene. Five accessions of sectionTimopheeviihad dominant alleles of bothVRN-A1andVRN-G1promoters. Both variants could contribute to the formation of spring growth habit.
因此,预测春天的增长来源的习惯was determined for 54 of 57 tested accessions, including those described previously as well as the novel disruption identified in the promoter or first intron sequences ofVRN-A1,VRN-B1和/或VRN-G1genes. The rest of the accessions, which includet . aethiopicumK-19059,T. turanicumK-31693 andT. turgidumcv. Blancal (K-20416), contained the recessiveVRN-A1andVRN-B1promoter sequences and an intact first intron of bothVRN-A1andVRN-B1genes. A cause for the spring growth habit in these accessions remains unknown.
Origin ofVRN-A1promoter variability in tetraploid wheat species
The variability identified in this study is probably the direct cause of the differences between spring and winter growth habit in tetraploid wheat species. To date, 20 different alleles of theVRN-A1promoter were identified in the genomes of di- and tetraploid species (Fig.3; Additional file2: Figure S1). Three recessive alleles differing by short deletions are presented in diploids, while the remaining four dominant alleles possess different substitutions, deletions and insertions compared to the recessive alleles. Three recessive and eleven dominant are presented among tetraploid accessions (Figs.3and4).
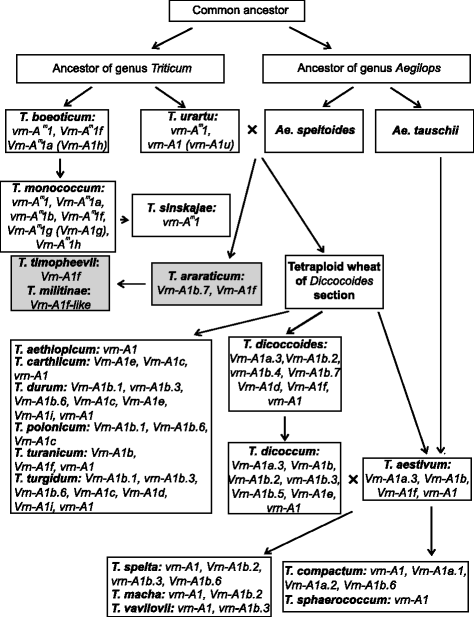
年代cheme ofTriticumandAegilopsgenera evolution (according to Goncharov [41], with additions). Different alleles ofVRN-A1gene among wheat species are presented in appropriate boxes next to the species names. SectionTimopheeeviiis presented in grey boxes, while sectionMonococcon, DicoccoidesandTriticumare in white boxes
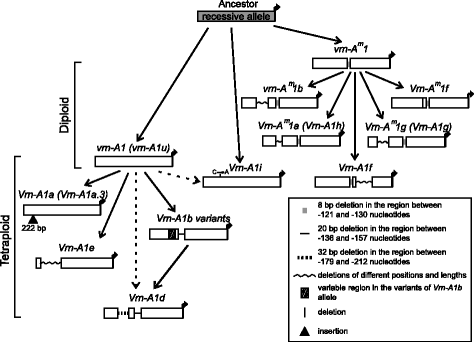
Analysis of the sequences allowed us to suggest the predicted pattern of evolution ofVRN-A1promoters. The recessive allelevrn-A1identified in tetraploid species was inherited from diploids, presumably fromT. urartu(Figs.3and4).
Dominant alleles虚拟现实n-A1a(虚拟现实n-A1a.3),虚拟现实n-A1e,虚拟现实n-A1iand variants of虚拟现实n-A1bfirst appeared in tetraploid species and apparently originated fromvrn-A1by way of short deletions in the case of虚拟现实n-A1band虚拟现实n-A1e, substitution in case of虚拟现实n-A1iand insertions for虚拟现实n-A1a.3(Fig.3). The variant of虚拟现实n-A1ballele except虚拟现实n-A1b.7and虚拟现实n-A1eare presented only in tetraploid species of theDicoccoidessection, and may originate from thevrn-A1allele after sections separation. Allele虚拟现实n-A1b.7is presented in bothDicoccoidesandTimopheeviisections and apparently originated in a common tetraploid wheat ancestor before sections separation (Fig.4). Among tetraploids the distribution of the虚拟现实n-A1a.3allele was restricted byT. dicoccumandT. dicoccoidesaccessions of theDicoccoidessection (Fig.4).
The dominant虚拟现实n-A1dallele is presented in bothDicoccoidesandTimopheeviisections, and may originate from one of the虚拟现实n-A1bvariants through extension of the deletion. Alternatively, the formation of two deletions in thevrn-A1allele could give the虚拟现实n-A1d(Fig.3). Allele虚拟现实n-A1dprobably originated only once in the ancestor of tetraploid wheat species and evidently was not inherited by hexaploid wheat species (Figs.3and4). Kato et al. [40] predicted that spring accessions ofT. dicoccoidesevolved from a winter forms as an adaptation to warmer conditions. However in the present investigation no correlation between the presence of虚拟现实n-A1dallele and particular environmental conditions of collection sites of studied accessions was identified (Table1).
The most interesting case is the虚拟现实n-A1fallele, which in comparison to other dominant alleles of tetraploid wheat, originated from the recessivevrn-Am1allele of a common ancestor of diploid wheat species (Fig.3).虚拟现实n-A1fallele is on a par with dominant alleles of diploids obtained by deletion in the recessivevrn-Am1allele promoter. To date, the虚拟现实n-A1fallele has been identified in diploids (T. monococcum,T. urartu,T. boeoticum), tetraploids (DicoccoidesandTimopheeviisections) and hexaploids (T. aestivum) (Fig.4) [16,17]。
Conclusions
The growth habit was investigated for 228 accessions of 12 tetraploid wheat species. The promoter and first intron regions ofVRN1genes were analyzed in 57 spring accessions of 10 tetraploid species. Comparative analysis revealed the novel allele ofVRN-A1(虚拟现实n-A1b.7) andVRN-B1(虚拟现实n-B1dic).虚拟现实n-A1dwas widely distributed across the accessions ofT. dicoccoides. In the genetic experiments the dominant mode of inheritance was shown for the虚拟现实n-A1dand虚拟现实n-A1calleles. It is assumed that the presence of虚拟现实n-A1dallele is associated with the formation of spring growth habit in the 11 accessions ofT. dicoccoides. Vrn-B1dicis a unique allele characterized by the unexpected high level of promoter sequence dissimilarity in comparison to thevrn-B1. This allele was identified in the only accession ofT. dicoccoides(IG46225) and further investigations are required to determine the role of this allele in the formation of spring growth habit. Novel allelic variants identified in the represent study provide a useful resource for fundamental investigations and could be used in agricultural production to expand the biodiversity of cultivated of wheat species. The summarization of the results regarding to theVRN1alleles identified to date in di- and polyploid wheat species allowed us to discuss the evolution of the alleles.
References
- 1.
Cockram J, Jones H, Leigh FJ, O’Sullivan D, Powell W, Laurie DA, Greenland AJ. Control of flowering time in temperate cereals: Genes, domestication, and sustainable productivity. J Exp Bot. 2007;58(6):1231–44.
- 2.
Distelfeld A, Li C, Dubcovsky J. Regulation of flowering in temperate cereals. Curr Opin Plant Biol. 2009;12(2):178–84.
- 3.
Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J. Positional cloning of the wheat vernalization geneVRN1. Proc Natl Acad Sci U S A. 2003;100:6263–8.
- 4.
Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J. The wheatVRN2gene is a flowering repressor down-regulated by vernalization. Science. 2004;303(5664):1640–4.
- 5.
Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J. The wheat and barley vernalization geneVRN3is an orthologue of FT. Proc Natl Acad Sci. 2006;103(51):19581–6.
- 6.
Kippes N, Debernardi JM, Vasquez-Gross HA, Akpinar BA, Budak H, Kato K, Chao S, Akhunov E, Dubcovsky J. Identification of theVERNALIZATION 4gene reveals the origin of spring growth habit in ancient wheats from South Asia. Proc Natl Acad Sci U S A. 2015;112(39):E5401–10.
- 7.
Trevaskis B, Hemming MN, Dennis ES, Peacock WJ. The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci. 2007;12(8):352–7.
- 8.
Fjellheim S, Boden S, Trevaskis B. The role of seasonal flowering responses in adaptation of grasses to temperate climates. Front Plant Sci. 2014;5:431.
- 9.
Law CN, Worland AJ, Giorgi B. The Genetic Control of Ear-Emergence Time by Chromosomes 5A an 5D of Wheat. Heredity (Edinb). 1976;36(1):49–58.
- 10.
Dubcovsky J, Lijavetzky D, Appendino L, Tranquilli G. Comparative RFLP mapping ofTriticum monococcumgenes controlling vernalization requirement. Theor Appl Genet. 1998;97(5–6):968–75.
- 11.
Barrett B, Bayram M, Kidwell K. Identifying AFLP and microsatellite markers for vernalization response gene虚拟现实n-B1in hexaploid wheat using reciprocal mapping populations. Plant Breed. 2002;121(5):400–6.
- 12.
Iwaki K, Nishida J, Yanagisawa T, Yoshida H, Kato K. Genetic analysis of虚拟现实n-B1for vernalization requirement by using linked dCAPS markers in bread wheat (Triticum aestivumL.). Theor Appl Genet. 2002;104(4):571–6.
- 13.
Danyluk J, Kane NA, Breton G, Limin AE, Fowler BD, Sarhan F. TaVRT-1, a Putative Transcription Factor Associated with Vegetative to Reproductive Transition in Cereals. Plant Physiol. 2015;132:1849–60.
- 14.
Murai K, Miyamae M, Kato H, Takumi S, Ogihara Y. WAP1, a WheatAPETALA1Homolog, Plays a Central Role in the Phase Transition from Vegetative to Reproductive Growth. Plant Cell Physiol. 2003;44(12):1255–65.
- 15.
Dubcovsky J, Loukoianov A, Fu D, Valarik M, Sanchez A, Yan L. Effect of photoperiod on the regulation of wheat vernalization genesVRN1andVRN2. Plant Mol Biol. 2006;60(4):469–80.
- 16.
Golovnina KA, Kondratenko EY、Goncharov Blinov AG)NP. Molecular characterization of vernalization loci VRN1 in wild and cultivated wheats. BMC Plant Biol. 2010;10:168.
- 17.
年代hcherban AB, Strygina KV, Salina EA.VRN-1gene- associated prerequisites of spring growth habit in wild tetraploid wheatT. dicoccoidesand the diploid A genome species. BMC Plant Biol. 2015;15(1):94.
- 18.
Pidal B, Yan L, Fu D, Zhang F, Tranquilli G, Dubcovsky J. The CArG-box located upstream from the transcriptional start of wheat vernalization geneVRN1is not necessary for the vernalization response. J Hered. 2009;100(3):355–64.
- 19.
Muterko A, Kalendar R, Salina E. Novel alleles of theVERNALIZATION1genes in wheat are associated with modulation of DNA curvature and flexibility in the promoter region. BMC Plant Biol. 2016;16 Suppl 1:65–81.
- 20.
Yan L, Helguera M, Kato K, Fukuyama S, Sherman J, Dubcovsky J. Allelic variation at theVRN-1promoter region in polyploid wheat. Theor Appl Genet. 2004;109(8):1677–86.
- 21.
Chu C, Tan C, Yu G, Zhong S, Xu SS, Yan L. A Novel Retrotransposon Inserted in the Dominant虚拟现实n-B1Allele Confers Spring Growth Habit in Tetraploid Wheat (Triticum turgidumL.). G3 (Bethesda). 2011;1(7):637–45.
- 22.
Fu D, Szucs P, Yan L, Helguera M, Skinner JS, von Zitzewitz J, Hayes PM, Dubcovsky J. Large deletions within the first intron inVRN-1are associated with spring growth habit in barley and wheat. Mol Genet Genomics. 2005;273(1):54–65.
- 23.
Ivaničová Z, Jakobson I, Reis D, Safar J, Milec Z, Abrouk M, Doležel J, Järve K, Valárik M. Characterization of new allele influencing flowering time in bread wheat introgressed fromTriticum militinae. N Biotechnol. 2016;33(5 Pt B):718–27.
- 24.
年代hcherban AB, Efremova TT, Salina EA. Identification of a new虚拟现实n-B1allele using two near-isogenic wheat lines with difference in heading time. Mol Breed. 2012;29(3):675–85.
- 25.
年代hcherban AB, Khlestkina EK, Efremova TT, Salina EA. The effect of two differentially expressed wheatVRN-B1alleles on the heading time is associated with structural variation in the first intron. Genetica. 2013;141(4):133–41.
- 26.
Milec Z, Sumikova T, Tomkova L, Pankova K. Distribution of different Vrn-B1 alleles in hexaploid spring wheat germplasm. Euphytica. 2013;192(3):371–8.
- 27.
年代antra DK, Santra M, Allan RE, Campbell KG, Kidwell KK. Genetic and Molecular Characterization of Vernalization Genes虚拟现实n-A1,虚拟现实n-B1, and虚拟现实n-D1in Spring Wheat Germplasm from the Pacific Northwest Region of the U.S.A. Plant Breed. 2009;128(6):576–84.
- 28.
Muterko A, Balashova I, Cockram J, Kalendar R, Sivolap Y. The New Wheat Vernalization Response Allele虚拟现实n-D1sis caused by DNA Transposon Insertion in the First Intron. Plant Mol Biol Report. 2015;33(2):294–303.
- 29.
Zhang X, Gao M, Wang S, Chen F, Cui D. Allelic variation at the vernalization and photoperiod sensitivity loci in Chinese winter wheat cultivars (Triticum aestivumL.). Front Plant Sci. 2015;6:470.
- 30.
Zhang J, Wang Y, Wu S, Yang J, Liu H, Zhou Y. A single nucleotide polymorphism at the虚拟现实n-D1promoter region in common wheat is associated with vernalization response. Theor Appl Genet. 2012;125(8):1697–704.
- 31.
Goncharov NP. Genetics of Growth Habit (Spring vs. Winter) in Tetraploid Wheats: Production and Analysis of Near-Isogenic Lines. Hereditas. 1999;130(2):125–30.
- 32.
Pugsley A. A genetic analysis of the spring-winter habit of growth in wheat. Aust J Agric Res. 1971;22:21.
- 33.
Goncharov NP. Response to vernalization in wheat: its quantitative or qualitative nature. Cereal Res Commun. 2004;32(3):323–30.
- 34.
Lu G, Moriyama EN. Vector NTI, a balanced all-in-one sequence analysis suite. Brief Bioinform. 2004;5(4):378–88.
- 35.
Okonechnikov K, Golosova O, Fursov M, Varlamov A, Vaskin Y, Efremov I, Grehov G, Kandrov D, Rasputin K, Syabro M, Tleukenov T. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics. 2012;28(8):1166–7.
- 36.
Goncharov NP. Genetic resources of wheat related species: the虚拟现实ngenes controlling growth habit (spring vs winter). Euphytica. 1998;100:371–6.
- 37.
Chhuneja P, Arora JK, Kaur P, Kaur S, Singh K. Characterization of wild emmer wheatTriticum dicoccoidesgermplasm for vernalization alleles. J Plant Biochem Biotechnol. 2015;24(2):249–53.
- 38.
Glover NM, Daron J, Pingault L, Vandepoele K, Paux E, Feuillet C, Choulet F. Small-scale gene duplications played a major role in the recent evolution of wheat chromosome 3B. Genome Biol. 2015;16:188.
- 39.
Airoldi CA, Davies B. Gene duplication and the evolution of plant MADS-box transcription factors. J Genet Genomics. 2012;39(4):157–65.
- 40.
Kato K, Mori Y, Beiles A, Nevo E. Geographical variation in heading traits in wild emmer wheat,Triticum dicoccoides. I. Variation in vernalization response and ecological differentiation. Theor Appl Genet. 1997;95:546–52.
- 41.
Goncharov NP. GenusTriticumL. taxonomy: The present and the future. Plant Syst Evol. 2011;295(1):1–11.
- 42.
Dorofeev VF, Filatenko AA, Migushova FF, Udachin RA, Jakubtsiner MM, Cultivated Flora of the USSR. Vol. 1. Pshenitsa (Wheat). Leningrad: Kolos; 1979 (In Russian).
Acknowledgments
We thank Ms. Carly Schramm, School of Biological Sciences, Flinders University (Australia), for critical review of our manuscript.
Declarations
This article has been published as part of BMC Plant Biology Volume 16 Supplement 3, 2016: Selected articles from BGRS\SB-2016: plant biology. The full contents of the supplement are available online at//www.cinefiend.com/articles/supplements/volume-16-supplement-3.
Funding
Funding for this work was provided by the RAS project number 21 (grant number: 0324-2015-0009) and the Russian Foundation for Basic Research (Grant No. 16-34-00688). Publication of this article has been funded by the Russian Foundation for Basic Research (Grant No. 16-34-00688).
Availability of data and material
TheVRN1gene sequences obtained in this study are available in GenBank, with the accession numbers KP063935-KP063977, KP063980-KP063992, KP063995-KP064001, KP064003-KP064012, KP260494, KP260496, KP260497, KR055675-KR055677, KR055679, KR055680, KR055683, KR055684, KR055686, KR055688, KR055694, KR055695, KR055697, KR055698.
Authors’ contributions
IK analyzed theVRN1genes sequences and wrote the manuscript. VV cloned and sequencedVRN1基因和导致了马的准备nuscript. EYaK carried out the greenhouse experiments. AB conceived and designed the molecular genetics experiments, revised the manuscript. NPG designed the greenhouse experiments, analyzed the results, contributed to the discussion and participated in preparing the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Author information
Affiliations
Corresponding author
Additional files
Additional file 2: Figure S1.
年代chematic representation ofVRN-A1(A),VRN-B1andVRN-G1(B) promoter region variability identified in di- and tetraploid wheat species. Insertions are indicated by black triangles, insertion lengths are marked under the black triangles. Deletions are indicated by dotted black lines. Hatched rectangles indicate variable region in the variants of虚拟现实n-A1ballele. Locations of the deletions and insertions are marked in base pair numbers upstream from the start codon, in accordance with the sequencevrn-A1u(GenBank Ac.No GQ451819). * - allele was identified by Golovnina et al. 2010.Figure S2.Geographical distribution of different variants ofVRN-A1gene from accessions of studied tetraploid wheat species. Abbrevations: a -虚拟现实n-A1a.3; b - variants of虚拟现实n-A1b; d -虚拟现实n-A1d; e -虚拟现实n-A1e; f -虚拟现实n-A1f; u -vrn-A1(vrn-A1u) allele. Numbers on the right of the graphic symbols match the numbers of studied wheat accessions which possess certain allelic variant.Figure S3.Distribution of heading time among studied accessions of ten tetraploid wheat species. Different species are marked by different colors.Figure S4.Distribution of heading time amongVRN-A1alleles identified in studied accessions of ten tetraploid wheat species. Different allelic variants are marked by different colors. (PDF 3302 kb)
Rights and permissions
Open AccessThis article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Konopatskaia, I., Vavilova, V., Kondratenko, E.Y.et al.VRN1genes variability in tetraploid wheat species with a spring growth habit.BMC Plant Biol16,244 (2016). https://doi.org/10.1186/s12870-016-0924-z
Published:
Keywords
- Evolution
- Growth habit
- Triticum
- vernalization
- VRN1gene
- Wheat