- Research article
- Open Access
- Published:
The expression of a tubby-like protein fromMalus domestica(MdTLP7) enhances abiotic stress tolerance inArabidopsis
BMC Plant Biologyvolume19, Article number:60(2019)
Abstract
Background
Tubby-like proteins (TLPs), characterized by a signature tubby domain, are widespread in plants and animals. To date, only plant TLPs involved in multifarious stress responses and male gametophyte development have been identified. However, studies on the molecular functions of plant TLPs are largely unknown.
Results
In this investigation, the roles of a TLP fromMalus domestica(MdTLP7) in response to abiotic stresses were characterized by expressing it inArabidopsis.The expression of wild-type full-lengthMdTLP7 (FL) significantly increased the stress tolerance ofArabidopsisseedlings to osmotic, salt, cold and heat stress, while the expression of truncatedMdTLP7 containing only the tubby domain (Tub) also showed some function. Located on a central α helix surrounded by 12 anti-parallel β strands in the tubby domain, the K190/R192site may be involved in fixation to the plasma membrane, as shown by 3D homology modelling with animal TLPs. This site might play a crucial role in anti-stress functions since site-directed mutagenesis ofMdTLP7 reduced stress tolerance. Subcellular localization showed thatMdTLP7 was mainly localized in the plasma membrane in plant cells, suggesting that it might participate in the transduction of stress signals.
Conclusions
The results of this study showed thatMdTLP7可以提高非生物压力tolerance not only in bacteria but also in plants. The K190/R192residues in the tubby domain were not only the plasma membrane binding site ofMdTLP7但在压力toleranc也发挥了关键作用e. These results may provide a basis for further exploring the mechanism of anti-stress functioning and downstream target genes of plant TLPs.
Background
The tubby-like proteins (TLPs) are widely distributed in the animal and plant kingdoms. There are 5 members of the TLP family in mice, 4 inHomo sapiens, 9 in apple and 11 inArabidopsis[1,2,3,4]。In mammals, many cellular functions of TLPs are involved in vesicular trafficking, the mediation of insulin signalling, gene transcription, G-protein signalling, and ribosomal RNA synthesis [5,6,7,8,9]。Although severalTLPs in plants have been identified, their roles are elusive. InArabidopsis,AtTLP3 andAtTLP9 are involved in ABA-dependent signalling during germination [3]。Several plantTLPs are upregulated under abiotic and biotic stress [4,10,11,12,13]。
Our previous study showed that aTLPgene fromMalus domestica(MdTLP7) was upregulated in the transcriptional profile of apple under cold stress [14]。Heterologous expression ofMdTLP7significantly increased the stress tolerance ofE. colicells against different abiotic stresses [15]。In contrast to animal TLPs,MdTLP7 contains a highly conserved F-box domain at its N-terminus in addition to the characteristic tubby domain [15]。In eukaryotes, F-box proteins participate in diverse cellular processes such as the response to stress, signal transduction, and the development of floral organs, primarily as a component of the Skp1-cullin-F-box (SCF) complex in protein ubiquitination [16,17,18,19,20]。
Subcellular localization of TLPs clarifies their functions. Many mammalian and plant TLPs are usually localized in the plasma membrane and nucleus [12,21]。As to the mechanism of TLP binding to the plasma membrane, a phosphatidylinositol-4,5-bisphosphate (PIP2) binding site involving two conserved residues in the tubby domain, lysine330(K330) and arginine332(R332), interacted directly with the membrane lipid of the plasma membrane, as demonstrated in two TLPs from mouse (TUB and TULP1) [22]。These two positively charged phosphate-coordinating residues (K330and R332) are highly conserved not only in the animal TLPs but also in quite a few plant TLPs. The corresponding plasma membrane binding site in the tubby domain ofAtTLP3 (K187/R189) also localizesAtTLP3 in the plasma membrane. Mutations of these conservative residues to other residues could disrupt the localization ofAtTLP3 to the plasma membrane [23]。PIP2affects cell signalling by binding target proteins or enzymes [24]。When plants suffer abiotic stress-inducing agents such as mannitol, NaCl and H2O2treatments,AtTLP3 can trigger signals by detaching from the plasma membrane and moving through the cytosol to the nucleus to regulate the expression of genes [12,23]。As bipartite transcription factors (TFs), mouse TLPs have DNA binding domains near their C-terminus and transcription modulation segments at their N-terminus [5]。TFs usually bind tocis-regulatory regions of a gene, which act as molecular switches controlling various biological processes including abiotic and biotic stress responses [25,26]。A chickpea TLP (CaTLP1) was demonstrated to be a putative TF presumably involved in multivariate stresses [11]。A rice TLP (OsTLP2) has been shown to bind to the promoter ofOsWRKY13to regulate disease resistance [10]。These reports suggest that TLPs might participate in stress signal transduction as TFs.
To determine the functions ofMdTLP7 in plants, full-length and truncatedMdTLP7 (without the F-box domain) were transformed intoArabidopsisin this study. The responses of transgenic plants to different stresses were assessed. Using 3D structure modelling and site-directed mutation experiments, we found a critical site in the tubby domain ofMdTLP7 for abiotic stress tolerance. These results open up new horizons for studying the mechanism of action of stress tolerance in plant TLPs and may promote the development of methods to improve plant stress resistance.
Results
Expression ofMdTLP7enhanced abiotic stress tolerance inArabidopsis
To examine the function ofMdTLP7in the plant stress response, the full-length cDNAs of wild-type full-lengthMdTLP7(FL) and truncatedMdTLP7with only the tubby domain (Tub) were transformed into wild-typeArabidopsis(WT) by the CaMV 35S promoter. More than 10 transgenic lines were identified by kanamycin testing and PCR amplification (T1 generation, Additional file1: Figure S1). Homozygous lines were identified by screening for non-segregation from each independent transformant (T3 generation). The 2 homozygous FL transgenic lines (named FL-1, FL-2) and 2 Tub transgenic lines (named Tub-1, Tub-2) with high expression were chosen for further analysis (Additional file2: Figure S2).
Under PEG and salt treatments, the growth of both transgenic lines, FL and Tub, was better than that of WT; little difference was found between the FL and Tub lines (Fig.1aandb). The results suggested that the expression of wild-typeMdTLP7 (FL) or truncatedMdTLP7 (Tub) enhanced tolerance to osmotic and salt stresses.
Phenotypes ofMdTLP7transgenic plants under abiotic stress.a20% PEG6000 treatment for 13 days,b250 mM NaCl treatment for 10 days,c45 °C treatment for 12 h and then normal growth conditions (22 °C) for 3 days, anddsuccessive pre-treatments at 5 °C to − 5 °C for 10 h followed by treatment at − 20 °C for 40 min and normal growth conditions (22 °C) for 3 days
To induce temperature stress, we placed seedlings in hot (45 °C) and cold (− 20 °C) environments. After treatment at 45 °C for 12 h, obvious differences were observed among the transgenic lines (FL and Tub) and between the transgenic lines and WTArabidopsis,and the growth of the Tub plants was better than that of the WT plants but worse than that of the FL plants. After 3 days of recovery, all of the WT plants were dead whereas 80% of the FL transgenic plants and 40% of the Tub transgenic plants survived (Fig.1c). Similar results after cold treatment were observed among the WT, FL and Tub lines. The seedlings were pre-treated at 5°Cto − 5 °C for 10 h and then subjected to − 20 °C for 40 min. All of the WT plants were dead whereas 70% of the FL plants and 30% of the Tub plants survived after 3 days of recovery (Fig.1d).
The results indicated that the expression ofMdTLP7 inArabidopsiscould enhance tolerance to several abiotic stressors inArabidopsis. TruncatedMdTLP7 (Tub) still retained almost complete function compared to that of wild-typeMdTLP7 (FL) under both PEG and salt stress. Regarding temperature stress, truncatedMdTLP7 (Tub) only had approximately half or less of the anti-stress function of wild-typeMdTLP7 (FL). The results suggested that the tubby domain ofMdTLP7 has key roles and that the F-box domain may have synergistic effects with the tubby domain in the stress response.
MdTLP7 is localized in the plasma membrane
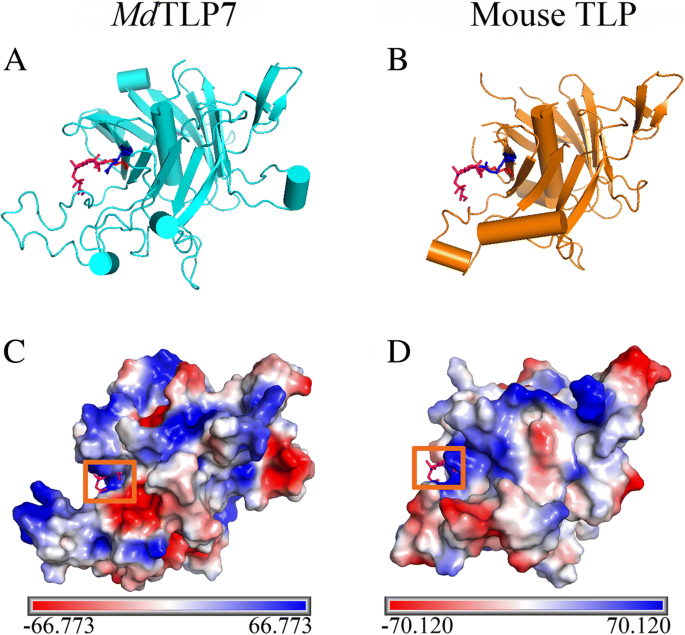
A 3D homology model of the tubby domains ofMdTLP7 was established using SWISS-MODEL that is highly consistent with the mouse TLP (TULP3) structure. Both contain a central α helix and a closed 12-stranded β barrel (Fig.2aandb). Mouse TLPs are tethered to the plasma membrane via a PIP2binding site (K330/R332) in the tubby domain [8]。The corresponding amino acid residues inMdTLP7, K190/R192, show high positional overlap with residues K330/R332of mouse TLP, the TLP plasma membrane binding site. Both grooves of the plasma membrane binding site inMdTLP7 and mouse TLP were positively charged as shown by electrostatic surface analysis (Fig.2candd). The results suggested thatMdTLP7 may also localize in the plasma membrane of plant cells, similar to mouse TLP in animal cells.
3D structures of the tubby domain ofMdTLP7 and mouse tubby protein.a-b3D homology model ofMdTLP7 (cyan) and mouse tubby protein (PDB identifier 1I7E; orange) showing K190(red) and R192(blue). IBS (pink), an analogue of PIP2, was used to show the conserved binding site.c-dElectrostatic surface of the tubby domain generated with the programme PyMOL. Electrostatic surface ofMdTLP7 with IBS modelled (c, orange frame) and mouse tubby protein-bound IBS (d, orange frame). The positive charges, negative charges and neutral charges are blue, red and white, respectively
To investigate whetherMdTLP7 localized in the plasma membrane, the sequence encodingMdTLP7 was fused to the sequence of GFP. As shown in Fig.3, the GFP signal emitted by the fusion protein was localized in the plasma membrane of tobacco leaf cells, while the GFP signal of the empty vector was localized in the cytoplasm and plasma membrane.
K190/R192in the tubby domain played crucial roles in stress tolerance
A PIP2analogue, L-α-glycerophospho-D-myoinositol-4,5-bisphosphate (IBS), was used to check changes in the charge of the corresponding plasma membrane binding site ofMdTLP7. When K190/R192were mutated to alanine residues, the electrostatic surface of the IBS-bound groove changed from positive to neutral charge, although the whole tertiary structure of the tubby domain was not significantly affected in 3D homology modelling (Additional file3: Figure S3 and Additional file4: Figure S4). Whether the positive charge of the active groove is indispensable for the function ofMdTLP7 needs to be identified.
To investigate the effects of this site on the anti-stress response, point mutants ofMdTLP (K190A, R192A or K190A/R192A) were constructed and transformed intoE. coli. A survival test on solid medium showed that the survival ratio of cells expressing wild-typeMdTLP7 was significantly higher than that ofMdTLP mutants under salt stress conditions (Additional file5: Figure S5).
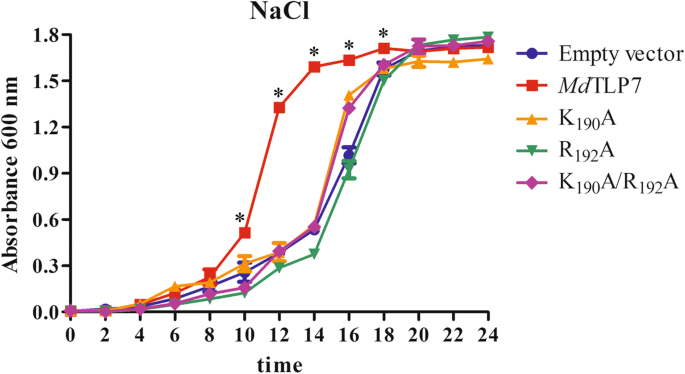
The growth curves of wild-typeMdTLP7 andMdTLP7 mutants in 500 mM NaCl-containing liquid medium were assayed to further ascertain the site conferring stress resistance toMdTLP7. The cells expressing wild-typeMdTLP7 entered the exponential growth stage after 10 h of cultivation, which was much earlier than the cells expressing mutantMdTLP7 (K190A, R192A or K190A/R192A). The three mutants entered the exponential growth stage at approximately 15 h, which was quite similar to the time frame of the empty vector (Table1and Fig.4). These results revealed that the plasma membrane biding site at K190/R192ofMdTLP7 played a key role in abiotic stress tolerance.
Discussion
肥胖的域名是签名守恒的域of the tubby-like family [9]。除了肥胖的领域,几乎所有的the plant TLPs have an F-box domain at their N-terminal regions [21]。MdTLP7 has two functional domains: an F-box domain at the N-terminus and a tubby domain at the C-terminus [4]。Some F-box domain-containing proteins are involved in the stress response by interacting with S-phase kinase association protein 1 to form an SCF complex and participating in the ubiquitin-proteasome system [27]。In our previous study, the F-box domain ofMdTLP7 was not essential for the anti-stress function of bacteria since truncation of the F-box did not significantly affect its anti-stress resistance [15]。In this study, only expressing the Tub domain ofMdTLP7 inArabidopsissignificantly increased tolerance to salt and PEG stress. Regarding cold and heat stress, the expression of full-lengthMdTLP7 was much better for tolerating cold and heat stress than the expression of truncatedMdTLP7. The presence of the conserved F-box domain inMdTLP7 might play a role in stress tolerance through the ubiquitin-proteasome pathway, which is a major posttranscriptional regulatory process related to redundant proteins in eukaryotes.
Boggon et al. determined that mouse TLPs were localized in the plasma membrane via a plasma membrane binding site (K330/R332) in a structure of the tubby domain bound to PIP2[5]。The 3D structure showed high positional overlap between K190/R192inMdTLP7 and K330/R332in mouse TLP (Fig.2). Therefore,MdTLP7 might also localize in the plasma membrane via K190/R192binding to PIP2. PIP2是一种膜磷脂影响特首ll signalling by binding target proteins to generate bioactive inositol phosphates [24]。In this investigation, we found thatMdTLP7 localized in the plasma membrane of tobacco cells, suggesting that it might participate in stress signal transduction (Fig.3). However, studies on the relationship between the plasma membrane binding site and the protein functions of plant TLP have not been reported. The survival ratios of cells expressingMdTLP7 point mutants (K190A and R192A) were slightly higher than that of cells expressing the empty vector under stress. However, the survival ratio of cells expressingMdTLP7 was significantly higher than that of cells expressingMdTLP7 mutants under salt stress condition (Additional file5: Figure S5). The growth curve assay also showed that the cells expressing wild-typeMdTLP7 entered the exponential growth stage much earlier than the cells expressing mutantMdTLP7 (Table1and Fig.4). The survival tests on solid and liquid medium both revealed that the plasma membrane binding site at K190/R192ofMdTLP7 played a key role in abiotic stress tolerance. The 3D homology model and electrostatic surface analysis ofMdTLP7 showed that point mutations at K190/R192did not change the 3D structure ofMdTLP7, but the PIP2binding groove changed from positively charged to neutral, indicating that the positive charge of the groove was important to the anti-stress function ofMdTLP7 (Additional file3: Figure S3 and Additional file4: Figure S4).
Conclusions
The results of this study showed thatMdTLP7可以提高非生物压力tolerance not only in bacteria but also inArabidopsis. The expression of full-length wild-typeMdTLP7 significantly increased the stress tolerance ofArabidopsisseedlings to osmotic, salt, cold and heat stress, while the expression of truncatedMdTLP7 including only the tubby domain also retained some function. The K190/R192residues ofMdTLP7 were found to be key amino acids involved in stress tolerance for the first time. In the future, we will further study the molecular mechanism ofMdTLP7 in abiotic stress tolerance.
Methods
Expression ofMdTLP7in transgenic plants
In this study,Arabidopsis thalianaecotype Columbia-0 (Col-0) was used as the experimental material.MdTLP7cDNA containing the complete open reading frame (ORF) and cDNA containing the tubby domain ofMdTLP7were amplified from theMdTLP7-pET30a recombinant vector, which was constructed in our previous study [15]。The full-lengthMdTLP7(FL) and tubby domain (Tub) cDNAs were inserted into the pBI121 vector, resulting in two recombinant vectors (pBI121-FL, pBI121-Tub). The pBI121-FL and pBI121-Tub recombinant vectors were integrated intoArabidopsisColumbia-0 byAgrobacterium tumefaciens(GV3101)介导的转换。花浸米ethod was used for this transformation [28]。Transformants (T1) were selected on Murashige and Skoog medium agar plates (50 μg/mL kanamycin). The presence and integrity of the FL or Tub cDNAs in transformants (T1) were further confirmed by PCR amplification. The primer sequences used in this experiment are listed in Additional file6S1:表。从公认的转基因种子plants were again selfed to obtain T3 progenies through T2 plants. The transgenic (T3 generation) and WTArabidopsisseeds were used for further experiments.
Quantitative real-time PCR (qRT-PCR) was performed to investigate the expression of FL and Tub inArabidopsis. Two FL transgenic plants (named FL-1 and FL-2), Tub transgenic plants (named Tub-1 and Tub-2) and wild-type (WT)Arabidopsisplants were randomly selected. FL-1 was used as a control, and theactingene was used as an internal control. The primer sequences used in this experiment are listed in Additional file6S1:表。The experiments were carried out at least three times under identical conditions. The relative expression levels were calculated using the 2-(ΔΔCt)method [29]。
Plant growth conditions and stress treatments
The WT, FL and Tub plants were grown in vermiculite at a constant temperature of 22 °C under 8 h illumination at 120 μmol m− 2 s− 1and a 16 h dark cycle. WT, FL and Tub plants were grown to 10 days of age. To induce osmotic stress, 10-day-old seedlings were irrigated with a 20% PEG6000 solution every day for up to 13 days. To induce salt stress, 10-day-old seedlings were irrigated with a 250 mM NaCl solution every day for up to 10 days.
FortyArabidopsisplants from four independent lines grown for 10 days at 22 °C were subjected to temperature stress treatment. To induce heat stress, 10-day-old seedlings were grown at 45 °C for 12 h and then returned to a growth chamber at 22 °C under 8 h illumination at 120 μmol m− 2 s− 1and a 16 h dark cycle for 3 days. To induce cold stress, 10-day-old seedlings were pre-treated at 5 °C to − 5 °C for 10 h successively and then subjected to − 20 °C for 40 min. After cold treatment, the seedlings were returned to a growth chamber at 22 °C under 8 h illumination at 120 μmol m− 2 s− 1and a 16 h dark cycle for 3 days before scoring the survival of the seedlings.
Subcellular localization ofMdTLP7
To determine the subcellular localization ofMdTLP7, theMdTLP7ORF without a termination codon was inserted upstream of the GFP gene. The 35S-GFP plasmid was used as a control. The subcellular localization experiment was carried out byAgrobacterium tumefaciensinfiltration into the leaves of tobacco as described by Jia et al. [30]。After 48 h of infiltration, a two photon laser confocal microscope (ZEISS, Germany) was used to observe the fluorescence in tobacco cells. Fluorescence was detected using a 505 to 550 nm bandpass filter for GFP. Image processing was performed with the Zeiss LSM image processing software (Zeiss).
Construction ofMdTLP7site-directed mutants
Three point mutations (K190A, R192A and K190A/R192A) were made by a QuikChange™ mutagenesis kit using the mutant primers listed in Additional file6S1:表。The threeMdTLP7point mutants were all inserted into the pET30a (+) expression plasmid (Novagen) and transformed intoE. colicells. All the constructed plasmids were sequenced by Sunny Biotechnology Company (Shanghai China) to confirm the correct sequences.
Survival test under salt stress on solid medium and in solution medium
The survival test and growth analysis under salt stress of theE. colidescribed above were carried out as described by Du et al. [15]。The cells were grown in LB solution medium to an OD600 of 0.4–0.6 at 37 °C, and expression of the recombinant proteins was induced for 2 h with 0.5 mM isopropyl-β-D-thiogalactopyranoside at 37 °C. For the survival test on solid medium, cultures were diluted to an OD600 of 0.6 and then diluted to 10− 2, 10− 3and 10− 4. Ten microliter cultures from each dilution (10− 2, 10− 3and 10− 4) were plated on solid LB medium supplemented with 500 mM NaCl and then incubated at 37 °C for 16 h. The colony number on each plate for the culture diluted to 10− 3was counted after incubation. Each experiment was repeated 3 times, and the data are presented as the mean with error bars indicating the standard deviation. The significance of the differences between groups was estimated using Student’st-test. A value ofP < 0.01 indicated a significant difference.
For the survival test in solution medium, cultures were diluted to an OD600 of 0.6 and 20 μl of the cultures were used to inoculate 10 ml LB solution containing a high concentration of 500 mM NaCl before incubation at 37 °C on a rotary shaker (150 rpm). The bacterial suspension was harvested every 2 h until 24 h had passed, and the OD600 was measured. Each experiment was repeated 3 times, and the data are presented as the mean with error bars indicating the standard deviation. All statistical analyses were performed using SPSS 19.0 (IBM, SPSS, Chicago, IL). Figures were made by GraphPad Prism 5 (GraphPad Software Inc., San Diego, CA, USA). Nonlinear regression analysis was fit using a logistic model (Y = A/ (1 + Be-kt)). The inflection point of the time ((lnB)/k) of each sample was calculated. The significance of the differences between groups was estimated using Student’st-test. A value ofP < 0.001 indicated a significant difference.
3D structure modelling
The 3D structural models ofMdTLP7 andMdTLP7 mutants were constructed using the protein structure homology-modelling server SWISS-MODEL [31,32,33]。The most suitable template structure searched by the SWISS-MODEL server was used. To compare the spatial position ofMdTLP7 bound to PIP2, a mouse brain tubby protein-bound PIP2structure (PDB identifier 1I7E) was used as a 3D model. Visualization of the protein molecules and vacuum electrostatics analysis was performed using the PyMOL Molecular Graphics System (Version 1.7.4.5,https://pymol.org/2/, Schrödinger, LLC).
Abbreviations
- 3D:
-
three-dimensional
- A:
-
alanine
- FL:
-
full-length ofMdTLP7
- IBS:
-
L-α-glycerophospho-D-myoinositol 4,5-bisphosphate
- K:
-
lysine
- ORF:
-
open reading frame
- PIP2:
-
phosphatidylinositol-4,5-bisphosphate
- qRT-PCR:
-
Quantitative real-time PCR
- R:
-
arginine
- SCF:
-
Skp1-cullin-F-box
- TF:
-
转录因子
- TLP:
-
tubby-like protein
- Tub:
-
tubby domain
- WT:
-
wild-type ofArabidopsis
References
- 1.
North MA, Naggert JK, Yan Y, Noben-Trauth K, Nishina PM. Molecular characterization of TUB, TULP1, and TULP2, members of the novel tubby gene family and their possible relation to ocular diseases. Proc Natl Acad Sci. 1997;94:3128–33.
- 2.
Nishina PM, North MA, Ikeda A, Yan Y, Naggert JK. Molecular characterization of a novel tubby gene family member, TULP3, in mouse and humans. Genomics. 1998;54:215–20.
- 3.
Lai CP, Lee CL, Chen PH, Wu SH, Yang CC, Shaw JF. Molecular analyses of the Arabidopsis TUBBY-like protein gene family. Plant Physiol. 2004;134:1586–97.
- 4.
Xu JN, Xing SS, Zhang ZR, Chen XS, Wang XY. Genome-wide identification and expression analysis of the tubby-like protein family in theMalus domesticagenome. Front Plant Sci. 2016;7:1693.
- 5.
Boggon TJ, Shan WS, Santagata S, Myers SC, Shapiro L. Implication of tubby proteins as transcription factors by structure-based functional analysis. Science. 1999;286:2119–25.
- 6.
Kapeller R, Moriarty A, Strauss A, Stubdal H, Theriault K, Siebert E, et al. Tyrosine phosphorylation of tub and its association with Src homology 2 domain-containing proteins implicate tub in intracellular signaling by insulin. J Biol Chem. 1999;274:24980–6.
- 7.
He W, Ikeda S, Bronson RT, Yan G, Nishina PM, North MA, Naggert JK. GFP-tagged expression and immunohistochemical studies to determine the subcellular localization of the tubby gene family members. Mol Brain Res. 2000;81:109–17.
- 8.
Santagata S, Boggon TJ, Baird CL, Gomez CA, Zhao J, Shan WS, Myszka DG, Shapiro L. G-protein signaling through tubby proteins. Science. 2001;292:2041–50.
- 9.
Ikeda A, Nishina PM, Naggert JK. The tubby-like proteins, a family with roles in neuronal development and function. J Cell Sci. 2002;115:9–14.
- 10.
Cai M, Qiu D, Yuan T, Ding X, Li H, Duan L, Xu C, Li X, Wang S. Identification of novel pathogen-responsive cis-elements and their binding proteins in the promoter of OsWRKY13, a gene regulating rice disease resistance. Plant Cell Environ. 2008;31:86–96.
- 11.
Wardhan V, Jahan K, Gupta S, Chennareddy S, Datta A, Chakraborty S, Chakraborty N. Overexpression of CaTLP1, a putative transcription factor in chickpea (Cicer arietinumL.), promotes stress tolerance. Plant Mol Biol. 2012;79:479–93.
- 12.
Bao Y, Song WM, Jin YL, Jiang CM, Yang Y, Li B, Huang WJ, Liu H, Zhang HX. Characterization ofArabidopsistubby-like proteins and redundant function of AtTLP3 and AtTLP9 in plant response to ABA and osmotic stress. Plant Mol Biol. 2014;86:471–83.
- 13.
Chen Y, Dai W, Sun B, Zhao Y, Ma Q. Genome-wide identification and comparative analysis of the TUBBY-like protein gene family in maize. Genes Genom. 2016;38:25–36.
- 14.
Du F, Xu JN, Li D, Wang XY. The identification of novel and differentially expressed apple-tree genes under low-temperature stress using high-throughput Illumina sequencing. Mol Biol Rep. 2015;42:569–80.
- 15.
Du F, Xu JN, Zhan CY, Yu ZB, Wang XY. An obesity-like gene MdTLP7 from apple (Malusxdomestica) enhances abiotic stress tolerance. Biochem Biophys Res Commun. 2014;445:394–7.
- 16.
Kiyosue T, Wada M. LKP1 (LOV kelch protein 1): a factor involved in the regulation of flowering time inarabidopsis. Plant J. 2000;23:807–15.
- 17.
Mas P, Kim WY, Somers DE, Kay SA. Targeted degradation of TOC1 by ZTL modulates circadian function inArabidopsis thaliana. Nature. 2003;426:567–70.
- 18.
Somers DE, Kim WY, Geng R. The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell. 2004;16:769–82.
- 19.
Baudry A, Ito S, Song YH, Strait AA, Kiba T, Lu S, Henriques R, et al. F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell. 2010;22:606–22.
- 20.
Takase T, Nishiyama Y, Tanihigashi H, Ogura Y, Miyazaki Y, Yamada Y, Kiyosue T. LOV KELCH PROTEIN2 and ZEITLUPE repress Arabidopsis photoperiodic flowering under non-inductive conditions, dependent on FLAVIN-BINDING KELCH REPEAT F-BOX1. Plant J. 2011;67:608–21.
- 21.
Mukhopadhyay S, Jackson PK. The tubby family proteins. Genome Biol. 2011;12:225.
- 22.
Mukhopadhyay S, Wen X, Chih B, Nelson CD, Lane WS, Scales SJ, et al. TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes Dev. 2010;24:2180–93.
- 23.
Reitz MU, Bissue JK, Zocher K, Attard A, Huckelhoven R, Becker K, et al. The subcellular localization of tubby-like proteins and participation in stress signaling and root colonization by the mutualistPiriformospora indica. Plant Physiol. 2012;160:349–64.
- 24.
Munnik T, Nielsen E. Green light for polyphosphoinositide signals in plants. Cur Opin Plant Biol. 2011;14:489–97.
- 25.
Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Cur Opin Plant Biol. 2003;6:410–7.
- 26.
Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005;10:88–94.
- 27.
Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol. 2009;10:385–97.
- 28.
Clough SJ, Bent AF. Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–43.
- 29.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–8.
- 30.
Jia H, Hao L, Guo X, Liu S, Yan Y, Guo X. A Raf-like MAPKKK gene, GhRaf19, negatively regulates tolerance to drought and salt and positively regulates resistance to cold stress by modulating reactive oxygen species in cotton. Plant Sci. 2016;252:267–81.
- 31.
Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201.
- 32.
Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27:343–50.
- 33.
Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–8.
Acknowledgements
Not Applicable.
Funding
This work was supported by National Natural Science Foundation of China (31872042). The Funding bodies were not involved in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Author information
Affiliations
Contributions
Designed the experiments: XW, SX and JX. Performed the experiments: SX, JX and CZ. Analyzed the data: XL, JX and SZ. Wrote the paper: XW, QS and JX. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
施普林格自然仍然neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Figure S1.PCR amplification of Kan-resistant seedlings. (1–10) PCR products of OEFLtransgenic lines, (11–20) PCR products of OETubtransgenic lines, M, DL2000 marker. (TIF 502 kb)
Additional file 2:
Figure S2.qRT-PCR analysis of the expression ofMdTLP7in the leaves of WT and transgenic plants. (TIF 1457 kb)
Additional file 3:
Figure S3.Homology model of the Tubby domain ofMdTLP7 and threeMdTLP7 point mutants. (A) Tubby domain ofMdTLP7. (B-D) ThreeMdTLP7 point mutants. K190is shown in red, R192in blue, and mutated amino acids in orange. (TIF 1046 kb)
Additional file 4:
Figure S4.Electrostatic surface of the tubby domain ofMdTLP7 and threeMdTLP7 point mutations. (A) Electrostatic surface of theMdTLP7 tubby protein. (B-D) Electrostatic surfaces of the point mutants. Blue indicates a positive charge, red indicates a negative charge and white indicates a neutral charge. The groove of the IBS-bound region is shown in the green frame. (TIF 2105 kb)
Additional file 5:
Figure S5.Survival test ofE. coliexpressingMdTLP7, empty vector or point mutants of under salt stress. (A) 10 μl cultures from 10− 2to 10− 4dilutions were spotted on LB plates treated with 0.5 M NaCl. (B) The colony numbers for the 10− 3dilutions appearing on each plate were counted. *p < 0.01. (TIF 13567 kb)
Additional file 6:
Table S1.Primers used in this experiment. (DOCX 12 kb)
Rights and permissions
Open AccessThis article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Xu, J., Xing, S., Sun, Q.et al.The expression of a tubby-like protein fromMalus domestica(MdTLP7) enhances abiotic stress tolerance inArabidopsis.BMC Plant Biol19,60 (2019). https://doi.org/10.1186/s12870-019-1662-9
Received:
Accepted:
Published:
DOI:https://doi.org/10.1186/s12870-019-1662-9
Keywords
- Tubby-like protein
- MdTLP7
- Functional site
- Stress tolerance
- Subcellular localization